Application of LC–MS/MS with ion mobility for chemical analysis of propolis extracts with antimicrobial potential Scientific paper
Main Article Content
Abstract
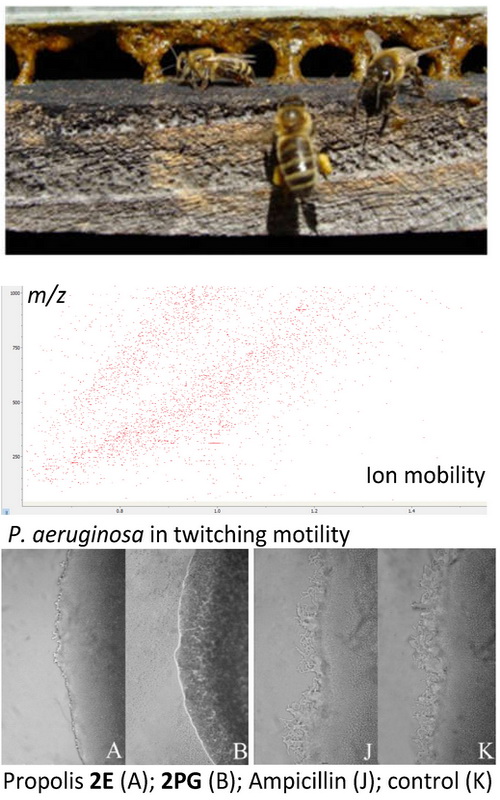
The objective of this study was to test four-dimensional LC-ESI-MS/MS chromatography in analysis of complex mixture such as ethanol extracts of different propolis samples. In total more than 1200 picks were identified and only for 185 literature conformation was found. The given data represent the result of tentative identification, and summarized results are given in the text. Comparing the samples, from different altitudes, 96 components were detected as characteristic in high altitude samples and 18 in samples collected at low altitudes. Antimicrobial activity of ethanol extracts of propolis (EEP) and propylene glycol extracts of propolis (PGEP) were carried out on S. aureus, B. cereus, M. flavus, L. monocytogenes, P. aeruginosa, S. typhimurium, E. coli and E. cloacae bacterial strains and compared with broad-spectrum antibiotics, streptomycin and ampicillin. Anti-quorum sensing activity was performed on P. aeruginosa by testing the effect of representative propolis extracts on biofilm formation, twitching and motility activity and production of pyocyanin. We demonstrated that the majority of explored propolis extracts have greater or equal minimal inhibitory concentration and minimum bactericidal concentration values compared to antibiotics, independently of the solvent used for the extraction. The samples collected from the highest altitude emerged as least active antimicrobial agents but with the greatest potential as anti-quorum sensing agents.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
A. K. Kuropatnicki, E. Szliszka, W. Krol, Evidence-Based Complement. Altern. Med. 2013 (2013) 1 (https://doi.org/10.1016/j.jpba.2016.12.003)
B. Anđelković, L. Vujisić, I. Vučković, V. Tešević, V. Vajs, D. Gođevac, J. Pharm. Biomed. Anal. 135 (2017) 217 (https://doi.org/10.1155/2013/964149)
Y. Ma, J.-X. Zhang, Y.-N. Liu, A. Ge, H. Gu, W.-J. Zha, X.-N. Zeng, M. Huang, Free Radic. Biol. Med. 101 (2016) 163 (https://doi.org/10.1016/j.freeradbiomed.2016.09.012)
F. Missima, A. A. da S. Filho, G. A. Nunes, P. C. P. Bueno, J. P. B. De Sousa, J. K. Bastos, J. M. Sforcin, J. Pharm. Pharmacol. 59 (2010) 463 (https://doi.org/10.1211/jpp.59.3.0017)
O. K. Mirzoeva, R. N. Grishanin, P. C. Calder, Microbiol. Res. 152 (1997) 239 (https://doi.org/10.1016/S0944-5013(97)80034-1)
K. Cui, W. Lu, L. Zhu, X. Shen, J. Huang, Biochem. Biophys. Res. Commun. 435 (2013) 289 (https://doi.org/10.1016/j.bbrc.2013.04.026)
L. Grenho, J. Barros, C. Ferreira, V. R. Santos, F. J. Monteiro, M. P. Ferraz, M. E. Cortes, Biomed. Mater. 10 (2015) 025004 (https://doi.org/10.1088/1748-6041/10/2/025004)
A. Meto, B. Colombari, A. Meto, G. Boaretto, D. Pinetti, L. Marchetti, S. Benvenuti, F. Pellati, E. Blasi, Microorganisms 8 (2020) 243 (https://doi.org/10.3390/microorganisms8020243)
F. Di Pierro, G. Derosa, P. Maffioli, A. Bertuccioli, S. Togni, A. Riva, P. Allegrini, A. Khan, S. Khan, B. A. Khan, N. Altaf, M. Zahid, I. D. Ujjan, R. Nigar, M. I. Khushk, et al., Int. J. Gen. Med. 14 (2021) 2359 (https://doi.org/10.2147/IJGM.S318720)
S. Huang, C.-P. Zhang, K. Wang, G. Li, F.-L. Hu, Molecules 19 (2014) 19610 (https://doi.org/10.3390/molecules191219610)
M. P. Popova, K. Graikou, I. Chinou, V. S. Bankova, J. Agric. Food Chem. 58 (2010) 3167 (https://doi.org/10.1021/jf903841k)
B. Trusheva, M. Popova, E. B. Koendhori, I. Tsvetkova, C. Naydenski, V. Bankova, Nat. Prod. Res. 25 (2011) 606 (https://doi.org/10.1080/14786419.2010.488235)
I. Dimkić, P. Ristivojević, T. Janakiev, T. Berić, J. Trifković, D. Milojković-Opsenica, S. Stanković, Ind. Crops Prod. 94 (2016) 856 (https://doi.org/10.1016/j.indcrop.2016.09.065)
A. C. H. F. Sawaya, I. Barbosa da Silva Cunha, M. C. Marcucci, Chem. Cent. J. 5 (2011) 27 (https://doi.org/10.1186/1752-153X-5-27)
D. G. Watson, E. Peyfoon, L. Zheng, D. Lu, V. Seidel, B. Johnston, J. A. Parkinson, J. Fearnley, Phytochem. Anal. 17 (2006) 323 (https://doi.org/10.1002/pca.921)
F. Pellati, G. Orlandini, D. Pinetti, S. Benvenuti, J. Pharm. Biomed. Anal. 55 (2011) 934 (https://doi.org/10.1016/j.jpba.2011.03.024)
C. G. Vasilopoulou, K. Sulek, A.-D. Brunner, N. S. Meitei, U. Schweiger-Hufnagel, S. W. Meyer, A. Barsch, M. Mann, F. Meier, Nat. Commun. 11 (2020) 331 (https://doi.org/10.1038/s41467-019-14044-x)
Bruker, Trapped Ion Mobility Spectrometry, https://www.bruker.com/en/products-and-solutions/mass-spectrometry/timstof.html (accessed August 4th, 2021)
T. Tsukatani, H. Suenaga, M. Shiga, K. Noguchi, M. Ishiyama, T. Ezoe, K. Matsumoto, J. Microbiol. Methods 90 (2012) 160 (https://doi.org/10.1016/j.mimet.2012.05.001)
F. Meier, Data Acquisition Methods for Next-Generation Mass Spectrometry-Based Proteomics, Der Ludwig-Maximilians-Universität München, 2018, German Network for Bioinformatics Infrastructure, https://msbi.ipb-halle.de/MetFrag/ (accessed August 4th, 2021)
K. Grecka, P. M. Kuś, P. Okińczyc, R. W. Worobo, J. Walkusz, P. Szweda, Molecules 24 (2019) 1732 (https://doi.org/10.3390/molecules24091732)
M. S. Regueira, S. R. Tintino, A. R. P. da Silva, M. do S. Costa, A. A. Boligon, E. F. F. Matias, V. de Queiroz Balbino, I. R. A. Menezes, H. D. Melo Coutinho, Food Chem. Toxicol. 107 (2017) 572 (https://doi.org/10.1016/j.fct.2017.03.052)
R. Wojtyczka, A. Dziedzic, D. Idzik, M. Kępa, R. Kubina, A. Kabała-Dzik, J. Smoleń-Dzirba, J. Stojko, M. Sajewicz, T. Wąsik, Molecules 18 (2013) 9623 (https://doi.org/10.3390/molecules18089623)
. D. Wojtyczka, M. Kępa, D. Idzik, R. Kubina, A. Kabała-Dzik, A. Dziedzic, T. J. Wąsik, Evidence-Based Complement. Altern. Med. 2013 (2013) 1 (https://doi.org/10.1155/2013/590703)
P. L. Gould, M. Goodman, P. A. Hanson Int J Pharmaceut. 19 (1984) 149 (https://doi.org/10.1016/0378-5173(84)90157-1)
A. E. Akca, G. Akca, F. T. Topçu, E. Macit, L. Pikdöken, I. Ş. Özgen, Biomed Res. Int. 2016 (2016) 1 (https://doi.org/10.1155/2016/3627463)
J. Bryan, P. Redden, C. Traba, Lett. Appl. Microbiol. 62 (2016) 192 (https://doi.org/10.1111/lam.12532)
D. S. de Oliveira Dembogurski, D. Silva Trentin, A. G. Boaretto, G. V. Rigo, R. C. da Silva, T. Tasca, A. J. Macedo, C. A. Carollo, D. B. Silva, Food Res. Int. 111 (2018) 661 (https://doi.org/10.1016/j.foodres.2018.05.033).