Curcumin as a potential multiple-target inhibitor against SARS-CoV-2 Infection: A detailed interaction study using quantum chemical calculations Scientific paper
Main Article Content
Abstract
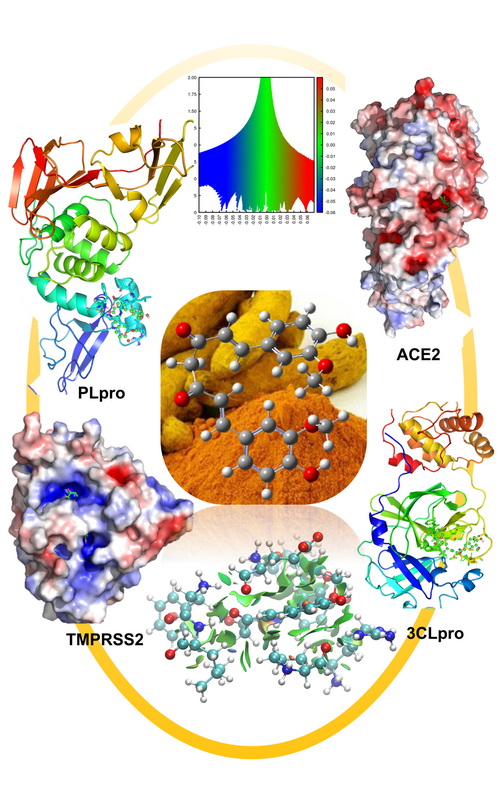
Curcumin is one of the important naturally occurring compounds having several medicinal properties such as: antiviral, antioxidant, antifibrotic, antineoplastic as well as anti-inflammatory. SARS-CoV-2 has emerged as infectious virus, which severely infected a large number of people all over the world. Many efforts have been made to prepare novel antiviral compound, but it is still challenging. Naturally occurring compound, curcumin, can be used as an alternative to antiviral compound against SARS-CoV-2. Its effect against SARS-CoV-2 is already highlighted in the literature. But the quantitative study of its interaction with various precursors of SARS-CoV-2 is not reported till date. This paper reports the interaction of curcumin with angiotensin-converting enzyme2, transmembrane serine protease 2, 3-chymotrypsin-like protease and papain-like protease through molecular docking and quantum chemistry calculations to achieve quantitative understanding of underlying interactions. Here the conformational flexibility of curcumin is also highlighted, which helps it to accommodate in the four different docking sites. The study has been performed using calculations of geometrical parameter, atomic charge, electron density, Laplacian of electron density, dipole moment and the energy gap between highest occupied and lowest unoccupied molecular orbitals. The non-covalent interaction (NCI) analysis is performed to visualize the weak interaction present in the active sites. Combinedly molecular docking and detailed quantum chemistry calculations revealed that curcumin can be adopted as a potential multiple-target inhibitor against SARS-CoV-2.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Department of Science and Technology, Ministry of Science and Technology, India
Grant numbers SRG/2019/002284
References
M. Cevik, M. Tate, O. Lloyd Maraolo, J. Schafers, A. Ho, Lancet Microbe 2 (2021) e13 (http://dx.doi.org/10.1016/S2666-5247(20)30172-5)
L. M. Mattio, G. Catinella, A. Pinto, S. Dallavalle, Eur. J. Med. Chem. 202 (2020) 112541 (http://dx.doi.org/10.1016/j.ejmech.2020.112541)
L. T. Lin, W. C. Hsu, C. C. Lin, J. Tradit. Complement. Med. 4 (2014) 24 (http://dx.doi.org/10.4103/2225-4110.124335)
F. A. C. Rocha, M. R. de Assis, Phytother. Res. 34 (2020) 2085 (http://dx.doi.org/10.1002/ptr.6745)
F. Zahedipour, S. A. Hosseini, T. Sathyapalan, M. Majeed, T. Jamialahmadi, K.
Al-Rasadi, M. Banach, A. Sahebkar, Phytother. Res. 34 (2020) 2911 (http://dx.doi.org/10.1002/ptr.6738)
V. K. Soni, A. Mehta, Y. K. Ratre, A. K. Tiwari, A. Amit, R. P. Singh, S. C. Sonkar, N. Chaturvedi, D. Shukla, N. K. Vishvakarma, Eur. J. Pharmacol. 886 (2020) 173551 (http://dx.doi.org/10.1016/j.ejphar.2020.173551)
F. Babaei, M. Nassiri-Asl, H. Hosseinzadeh, Food Sci. Nutr. 8 (2020) 5215 (http://dx.doi.org/10.1002/fsn3.1858)
R. K. Thimmulappa, K. K. Mudnakudu-Nagaraju, C. Shivamallu, K. J. T. Subramaniam, A. Radhakrishnan, S. Bhojraj, G. Kuppusamy, Heliyon 7 (2021) e06350 (http://dx.doi.org/10.1016/j.heliyon.2021.e06350)
A. Saeedi-Boroujeni, M. R. Mahmoudian-Sani, M. Bahadoram, A. Alghasi, Basic Clin. Pharmacol. Toxicol. 128 (2021) 37 (http://dx.doi.org/10.1111/bcpt.13503)
N. Chainani-Wu, J. Altern. Complement. Med. 9 (2003) 161 (http://dx.doi.org/10.1089/107555303321223035)
Y. Huang, C. Yang, X. F. Xu, W. Xu, S. W. Liu, Acta Pharmacol. Sin. 41 (2020) 1141 (http://dx.doi.org/10.1038/s41401-020-0485-4)
H. Zhang, J. M. Penninger, Y. Li, N. Zhong, A. S. Slutsky, Intensive Care Med. 46 (2020) 586 (http://dx.doi.org/10.1007/s00134-020-05985-9)
A. Domling, L. Gao, Chem 6 (2020) 1283 (http://dx.doi.org/10.1016/j.chempr.2020.04.023)
M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Kruger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Muller, C. Drosten, S. Pohlmann, Cell 181 (2020) 271 (http://dx.doi.org/10.1016/j.cell.2020.02.052)
A. B. Jena, N. Kanungo, V. Nayak, G. B. N. Chainy, J. Dandapat, Sci. Rep. 11 (2021) 2043 (http://dx.doi.org/10.1038/s41598-021-81462-7)
A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire, D. Veesler, Cell 181 (2020) 281 (http://dx.doi.org/10.1016/j.cell.2020.02.058)
N. Barretto, D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, S. C. Baker, J. Virol. 79 (2005) 15189 (http://dx.doi.org/10.1128/jvi.79.24.15189-15198.2005)
M. L. DeDiego, E. Alvarez, F. Almazán, M. T. Rejas, E. Lamirande, A. Roberts, W.-J. Shieh, S. R. Zaki, K. Subbarao, L. Enjuanes, J. Virol. 81 (2007) 1701 (http://dx.doi.org/10.1128/JVI.01467-06)
L. Kuo, P. S. Masters, J. Virol. 77 (2003) 4597 (http://dx.doi.org/10.1128/JVI.77.8.4597-4608.2003)
J. Ortego, J. E. Ceriani, C. Patino, J. Plana, L. Enjuanes, Virology 368 (2007) 296 (http://dx.doi.org/10.1016/j.virol.2007.05.032)
X. Y. Meng, H. X. Zhang, M. Mezei, M. Cui, Curr. Comput. Aided Drug Des. 7 (2011) 146 (http://dx.doi.org/10.2174/157340911795677602)
G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, A. J. Olson, J. Comput. Chem. 30 (2009) 2785 (http://dx.doi.org/10.1002/jcc.21256)
M. F. Sanner, J. Mol. Graph. Model. 17 (1999) 57 (http://dx.doi.org/10.1016/S1093-3263(99)99999-0)
R. A. Laskowski, M. B. Swindells, J. Chem. Inf. Model. 51 (2011) 2778 (http://dx.doi.org/10.1021/ci200227u)
A. C. Wallace, R. A. Laskowski, J. M. Thornton, Protein Eng. 8 (1995) 127 (http://dx.doi.org/10.1093/protein/8.2.127)
M. F. Adasme, K. L. Linnemann, S. N. Bolz, F. Kaiser, S. Salentin, V. J. Haupt, M. Schroeder, Nucleic Acids Res. 49 (2021) W530 (http://dx.doi.org/10.1093/nar/gkab294)
H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, P. E. Bourne, Nucleic Acids Res. 28 (2000) 235 (http://dx.doi.org/10.1093/nar/28.1.235)
M. D. Wodrich, C. Corminboeuf, P. v. R. Schleyer, Org. Lett. 8 (2006) 3631 (http://dx.doi.org/10.1021/ol061016i)
F. Weigend, R. Ahlrichs, Phys. Chem. Chem. Phys. 7 (2005) 3297 (http://dx.doi.org/10.1039/B508541A)
F. Weigend, Phys. Chem. Chem. Phys. 8 (2006) 1057 (http://dx.doi.org/10.1039/B515623H)
X. Xu, D. G. Truhlar, J. Chem. Theory Comput. 7 (2011) 2766 (http://dx.doi.org/10.1021/ct200234r)
J. Zheng, X. Xu, D. G. Truhlar, Theor. Chem. Acc. 128 (2011) 295 (http://dx.doi.org/10.1007/s00214-010-0846-z)
F. Furche, R. Ahlrichs, C. Hättig, W. Klopper, M. Sierka, F. Weigend, Wiley Interdiscip. Rev. Comput. Mol. Sci. 4 (2014) 91 (http://dx.doi.org/10.1002/wcms.1162)
C. Steffen, K. Thomas, U. Huniar, A. Hellweg, O. Rubner, A. Schroer, J. Comput. Chem. 31 (2010) 2967 (http://dx.doi.org/10.1002/jcc.21576)
E. R. Johnson, S. Keinan, P. Mori-Sanchez, J. Contreras-Garcia, A. J. Cohen, W. Yang, J. Am. Chem. Soc. 132 (2010) 6498 (http://dx.doi.org/10.1021/ja100936w).