Post-TRIzol protein extraction from peripheral blood mononuclear cells Scientific paper
Main Article Content
Abstract
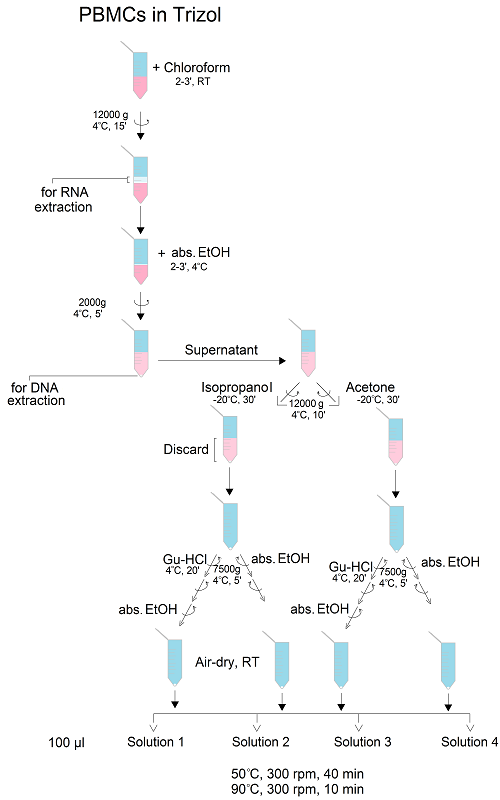
After sample processing for RNA and DNA analysis, the leftover protein pellets are usually discarded due to the limited efficiency of pellet reconstitution/solubilisation. As the pelleted proteins are tightly packed, they are most often solubilised using chaotropic agents (e.g., guanidine hydrochloride or urea), detergents (e.g., SDS), salts (NaCl) or basic buffer (Tris). The aim of this study was to define and optimise the procedure for the efficient extraction of proteins from human peripheral blood mononuclear cells (PBMCs), obtained by a single blood draw and lysed in TRIzol reagent, by varying experimental conditions in terms of protein precipitation solvent (isopropanol or acetone), washing (with or without guanidine hydrochloride) and solubilisation solution (containing SDS, NaCl, urea and/or Tris). We evaluated the efficacy of the final, optimised protocol to solubilise both small cytoplasmic and larger transmembrane proteins, and the compatibility with methods employed for the subsequent analysis of protein posttranslational modifications, such as glycosylation. The optimised protocol for the extraction and isolation of post-TRIzol leftover proteins from PBMCs can be defined as follows: protein precipitation from the organic phase with ice-cold acetone, pellet washing with absolute ethanol and solubilisation in 1 % SDS, employing 20 min heating at 50 °C and vortexing.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-47/2023-01/200019
References
W. Mathieson, G. A Thomas, Anal. Biochem. 433 (2013) 10 (https://dx.doi.org/10.1016/j.ab.2012.10.006)
P. Sen, E. Kemppainen, M. Orešič, Front. Mol. Biosci. 4 (2018) (https://dx.doi.org/10.3389/fmolb.2017.00096)
H. Riol, B. Jeune, A. Moskovic, L. Bathum, E. Wang, Anal. Biochem. 275 (1999) 192 (https://dx.doi.org/10.1006/abio.1999.4328)
A. M. Kopec, P. D. Rivera, M. J. Lacagnina, R. Hanamsagar, S. D. Bilbo, J. Neurosci. Methods 280 (2017) 64 (https://dx.doi.org/10.1016/j.jneumeth.2017.02.002)
R. B. Braakman, K. Bezstarosti, A. M. Sieuwerts, V. de Weerd, A. M. van Galen, C. Stingl, T. M. Luider, M. A. Timmermans, M. Smid, J. W. Martens, J. A. Foekens, J. A. Demmers, A. Umar, J. Proteome Res. 14 (2015) 1627 (https://dx.doi.org/10.1021/acs.jproteome.5b00046)
TRIsure™ Product Specifications, https://www.bioline.com/mwdownloads/download/link/id/953/trisure_product_manual.pdf accessed 25 July, 2023
C. Arndt, S. Koristka, A. Feldmann, R. Bergmann, M. Bachmann, Methods Mol. Biol. 1853 (2018) 27 (https://dx.doi.org/10.1007/978-1-4939-8745-04)
A. B. Hummon, S. R. Lim, M. J. Difilippantonio, T. Ried, Biotechniques 42 (2007) 467 (https://dx.doi.org/10.2144/000112401)
A. E. Simões, D. M. Pereira, J. D. Amaral, A. F. Nunes, S. E. Gomes, P. M. Rod¬rigues, A. C. Lo, R. D'Hooge, C. J. Steer, S. N .Thibodeau, P. M.Borralho, C. M. P. Rodrigues, BMC Genomics 14 (2013) 181 (https://dx.doi.org/10.1186/1471-2164-14-181)
A. P. Joy, D. C. Ayre, I. C. Chute, A. P. Beauregard, G. Wajnberg, A. Ghosh, S. M. Lewis, R. J. Ouellette, D. A. Barnett, J. Extracell. Vesicles 7 (2018) 1438727 (https://dx.doi.org/10.1080/20013078.2018.1438727)
Y. Wen, I. J. Vechetti, T. R. Valentino, J. J. McCarthy, Biotechniques 69 (2020) 264 (https://dx.doi.org/10.2144/btn-2020-0083)
C. Young, P. Truman, Anal Biochem. 421 (2012) 330 (https://doi.org/10.1016/j.ab.2011.10.045)
C. Pop, S. Ameling, D. Vishnu, F. Loghin, U. Völker, E. Hammer, JIOMICS 5 (2015) 58 (https://dx.doi.org/10.5584/jiomics.v5i1.185)
L. Hua, R. Zhou, D. Thirumalai, B.J. Berne, Proc. Natl. Acad. Sci. USA 105 (2008) 16928 (https://doi.org/10.1073/pnas.0808427105)
L. Kollipara, R.P. Zahedi, Proteomics 13 (2013) 941 (https://dx.doi.org/10.1002/pmic.201200452)
B. Schlager, A. Straessle, E. Hafen, BMC Biotechnol. 12 (2012) (https://dx.doi.org/10.1186/1472-6750-12-95).