Theoretical evaluation of pectin therapeutic potential in relation to degree of methylation Scientific paper
Main Article Content
Abstract
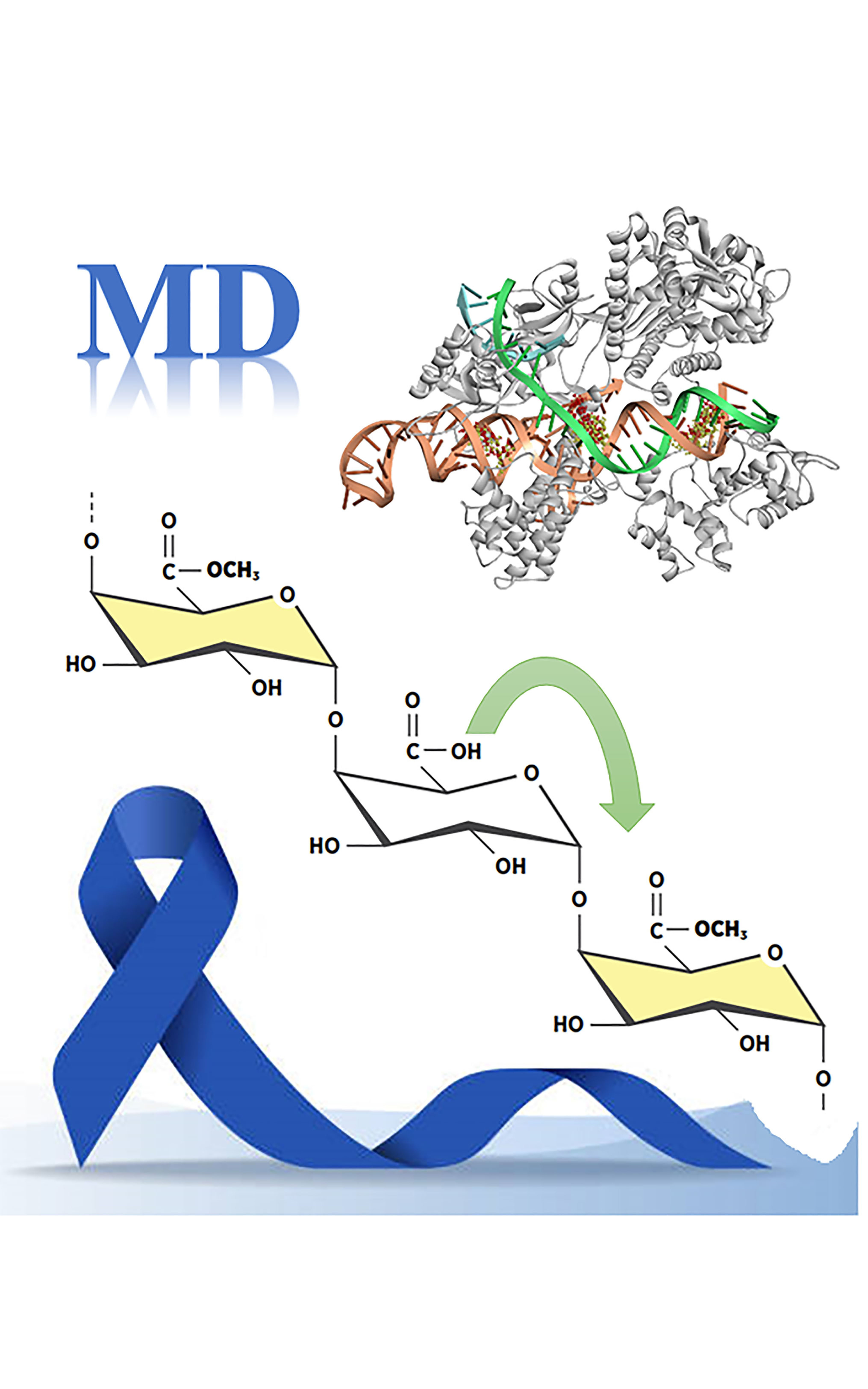
Pectin is the focus of scientific interest due to both its physicochemical and biochemical properties, as well as its non-toxic nature. Methylation of pectin is a natural process that exists as part of the cell wall defence system against various pathogens. In this study the docking analysis was conducted to predict if methylation o affects the anticancer and antimicrobial properties of pectin and twhat extent. Four pectin derivatives with varying degrees of methylation and two sets of biomolecules were used. The first set included enzymes responsible for anticancer activity (HMGR, the AGE Receptors, tumour protein p53, and Oncogenic Phosphatase SHP2), while the second set included those for antimicrobial activity (Salmonella Typhi TtsA, Pseudomonas aeruginosa Earp, Streptococcus mutans MetE, and Staphylococcus aureus Cas9). The results indicated that the degree of methylation does not play a decisive role in the mentioned activities. because all bind to the same sites with similar binding energies. Additionally, it was shown that pectin derivatives have a higher binding affinity towards DNA than towards enzymes. Only the fully methylated derivative exhibited different behaviour, binding to a different binding site in the case of Streptococcus mutans MetE.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministry of Scientific and Technological Development, Higher Education and Information Society,Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-66/2024-03/200026;451-03-66/2024-03/ 200162
References
A. Dambuza, P. Rungqu, A. Omowunmi Oyedeji, G. Miya, A. Oluwabunmi Oriola, Y. Yiseyon Sunday Hosu, O. Oyehan Oyedeji, Molecules 29 (2024) 896 (https://doi.org/10.3390/molecules29040896)
N. Koropatkin, E. Cameron, E. Martens, Nat. Rev. Microbiol. 10 (2012) 323 (https://doi.org/10.1038/nrmicro2746)
K. Shinohara, Z. Ohashi, K. Kawasumi, A. Terada, T. Fujisawa, Anaerobe 16 (2010) 410 (https://doi.org/10.1016/j.anaerobe.2010.03.005)
R. Ciriminna, A. Fidalgo, F. Meneguzzo, A. Presentato, A. Scurria, D. Nuzzo, R. Alduina, L. M. Ilharco, M. Pagliaro, ChemMedChem 15 (2020) 2228 (https://doi.org/10.1002/cmdc.202000518)
M. C. Wu, H. C. Li, P. H. Wu, P. H. Huang, Y. T. Wang, J. Food Sci. 79 (2014) 1541 (https://doi.org/10.1111/1750-3841.12526)
E. Calce, E. Mignogna, V. Bugatti, M. Galdiero, V. Vittoria, S. De Luca, Int. J. Biol. Macromol. 68 (2014) 28 (https://doi.org/10.1016/j.ijbiomac.2014.04.011)
T. B. Emran, F. Islam, S. Mitra, S. Paul, N. Nat, Z. Khan, R. Das, D. Chandran, R. Sharma, C. M. Gonçalves Lima, A. A. Al Awadh, I. A. Almazni, A. H. Alhasaniah, R. P. F. Guinéet, Molecules 27 (2022) 7405 (https://doi.org/10.3390/molecules27217405)
V. V. Glinsky, Carbohydr. Res. 344 (2009) 1788 (https://doi.org/10.1016/j.carres.2008.08.038)
E. A. Almeida, S. P. Facchi, A. F. Martins, S. Nocchi, I. T. A. Schuquel, C. V. Nakamura, A. F. Rubira, E. C. Muniz, Carbohydr. Polym. 115 (2015) 139 (https://doi.org/10.1016/j.carbpol.2014.08.085)
E. S. Istvan, M. Palnitkar, S. K. Buchanan, J. Deisenhofer, EMBO J. 19 (2000) 819 (https://doi.org/10.1093/emboj/19.5.819)
N. Kozlyuk, B. A. Gilston, L. E. Salay, R. D. Gogliotti, P. P. Christov, K. Kim, M. Ovee, A. G. Waterson, W. J. Chazin, Proteins 89 (2021) 1399 (https://doi.org/10.1002/prot.26162)
J. R. LaRochelle, M. Fodor, X. Xu, I. Durzynska, L. Fan, T. Stams, H. M. Chan, M. J. LaMarche, R. Chopra, P. Wang, P. D. Fortin, M. G. Acker, S. C. Blacklow, Biochemistry 55 (2016) 2269 (https://doi.org/10.1021/acs.biochem.5b01287)
M. R. Bauer, R. N. Jones, R. K. Tareque, B. Springett, F. A. Dingler, L. Verduci, K. J. Patel, A. R. Fersht, A. C. Joerger, J. Spencer, Future Med. Chem. 11 (2019) 2491 (https://doi.org/10.4155/fmc-2019-0181)
He, N. Liu, F. Li, X. Jia, H. Peng, Y.Liu, Y.Xiao , J. Bacteriol. 201 (2019) 1 (https://doi.org/10.1128%2FJB.00128-19)
T. Geiger, M. Lara-Tejero, Y. Xiong, J. E. Galán, eLife 9 (2020) e53473 (https://doi.org/10.7554/eLife.53473)
H. Nishimasu, L. Cong, W. X. Yan, F. A. Ran, B. Zetsche, Y.Li, A.Kurabayashi, R. Ishitani, F. Zhang, O. Nureki, Cell 162 (2015) 1113 (https://doi.org/10.1016/j.cell.2015.08.007)
T.-M. Fu, J. Almqvist, Y.-H. Liang, L. Li, Y. Huang, X.-D- Su, J. Mol. Biol. 412 (2011) 688 (https://doi.org/10.1016/j.jmb.2011.08.005)
G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, A. J. Olson, J. Comput. Chem. 30 (2009) 2785 (https://doi.org/10.1002/jcc.21256)
D. Biovia, H. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. Bhat, T. J. T. J. o. C. P. Richmond, Dassault Systèmes BIOVIA, Discovery Studio Visualizer, v. 17.2.0.16349, San Diego: Dassault Systèmes, 2016, 10 (2000) 0021-9991.
J. Martinov, M. Krstić, S. Spasić, S. Miletić, J. Stefanović-Kojić, A. Nikolić-Kokić, D. Blagojević, I. Spasojević, M. B. Spasić, Food Res. Int. 100 (2017) 132 (https://doi.org/10.1016/j.foodres.2017.08.040)
H. S. Park, J. S. Choi, K. H. Kim, Nutr. Res. 20 (2000) 1783 (https://doi.org/10.1016/S0271-5317(00)00269-4)
A. Markowska, M. Antoszczak, J. Markowska, A. Huczyński, Pharmaceuticals 13 (2020) 422 (https://doi.org/10.3390/ph13120422)
S. Bongarzone, V. Savickas, F. Luzi, A. D. Gee, J. Med. Chem. 60 (2017) 7213 (https://doi.org/10.1021/acs.jmedchem.7b00058)
Z. Li, B. Zhou, T. Zheng, C. Zhao, Y. Gao, W. Wu, Y. Fan, X. Wang, M. Qiu, J. Fan, Foods 12 (2023) 423 (https://doi.org/10.3390/foods12020423)
H. Yan, X. Zhang, L. Yang, Y. Shen, L. Liu, Food Chem. 398 (2023) 133886 (https://doi.org/10.1016/j.foodchem.2022.133886)
T. B. Emran, F. Islam, S. Mitra, S. Paul, N. Nath, Z. Khan, R. Das, D. Chandran, R. Sharma, C. M. G. Lima, A. A. Al Awadh, I. A. Almazni, A. H. Alhasaniah, R. P. F. Guiné, Molecules 27 (2022) 7405 (https://doi.org/10.3390/molecules27217405)
L. Delphi, H. Sepehri, Biomed. Pharmacother. 84 (2016) 637 (https://doi.org/10.1016/j.biopha.2016.09.080)
J. Zhang, F. Zhang, R. Niua, J. Cell. Mol. Med. 19 (2015) 2075 (https://doi.org/10.1111/jcmm.12618)
J. P. Skittrall, D. Levy, C. Obichukwu, A.Gentle , M. A. Chattaway , D. Hayns , C. Etheridge, C. M. Parry, V. Wong, J. Whitehorn, Clin. Infect. Pract. 10 (2021) 100069 (https://doi.org/10.1016/j.clinpr.2021.100069)
C. He, N. Liu, F. Li, X. Jia, H. Peng, Y. Liu, Y. Xiao, J. Bacteriol. 201 (2019) (https://doi.org/10.1128/JB.00128-19)
T.-M. Fu, J. Almqvist, Y.-H. Liang, L. Li, Y. Huang, X. D. Su, J. Mol. Biol. 412 (2011) 688 (https://doi.org/10.1016/j.jmb.2011.08.005)
W. Chen, Y. Zhang, W.S. Yeo, T. Bae, Q. Ji, J. Am. Chem. Soc. 139 (2017) 3790 (https://doi.org/10.1021/jacs.6b13317)
G. Hamion, W. Aucher, A. Mercier, F. Tewes, M. Menard, J. Bertaux, M. Girardot, C. Imbert, Int. J. Antimicrob. Agents 63 (2024) 107166 (https://doi.org/10.1016/j.ijantimicag.2024.107166).