Investigation of the bioactivities of Saponaria mesogitana methanolic extract along with its phytochemical composition Scientific paper
Main Article Content
Abstract
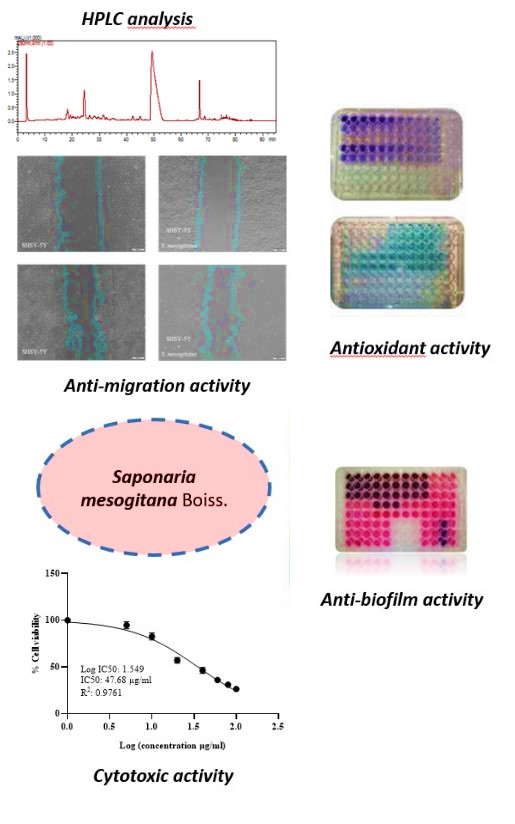
Saponaria species are known to contain saponins which have a wide variety of biological activities. But up to now, the phenolic compounds of Saponaria mesogitana have not been clarified. Therefore, this study aimed to determine the phenolic composition and some biological activities of S. mesogitana for the first time. The antioxidant activities of the methanol and water extracts were assessed using the DPPH, FRAP and β-carotene/linoleic acid assays, while the total secondary metabolite content, including phenolics, flavonoids and saponins, was also determined for both extracts. Based on the antioxidant activity and total phenolic and flavonoid contents, further HPLC analysis, as well as anticancer and antimicrobial activity experiments, were conducted using the methanol extract. The anticancer potential was assessed using the MTT assay and wound healing migration test, while antibacterial activity was evaluated through disc diffusion and MIC assays. Additionally, the antibiofilm properties of the extract were examined using the crystal violet method. The methanolic extract showed high antioxidant activity, while caffeic acid and epicatechin were characterized as major phenolic compounds by HPLC. S. mesogitana inhibited not only bacterial growth but also the levels of migration of SHSY-5Y cancer cells. These findings indicate that S. mesogitana possesses potent antioxidant, anticancer, antimicrobial and antibiofilm activities associated with its bioactive phenolic constituents.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
A. Akgul, A. Akgul, S. G. Senol, H. Yildirim, O. Secmen, Y. Dogan, J. Ethnobiol. Ethnomed. 14 (2018) 12 (https://doi.org/10.1186/s13002-017-0201-8)
M. Selseleh, S. N. Ebrahimi, A. Aliahmadi, A. Sonboli, M. H. Mirjalili, Ind. Crop Prod. 153 (2020) 112609 (https://doi.org/10.1016/j.indcrop.2020.112609)
M. J. Christenhusz, J. W. Byng, Phytotaxa 261 (2016) 201 (https://doi.org/10.11646/phytotaxa.261.3.1)
A. Dashti, M. Assadi, F. Sharifnia, Iran J. Bot. 20 (2014) 146 (https://doi.org/10.22092/IJB.2014.11008)
R. Pavela, Plant Protect. Sci. 52 (2016) 54 (https://doi.org/10.17221/62/2015-PPS)
G. B. Endonova, T. P. Antsupova, S. D. Zhamsaranova, D. V. Lygdenov, Biosci. Biotech. Res. Asia 12 (2015) 2017 (http://dx.doi.org/10.13005/bbra/1869)
M. Sengul, S. Ercişli, H. Yildiz, N. Gungor, A. Kavaz, B. Çetin, Iran. J. Pharm. Res. 10 (2011) 49 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3869595/)
J. J. Balsevich, I. Ramirez-Erosa, R. A. Hickie, D. M. Dunlop, G. G. Bishop, L. K Deibert, Fitoterapia 83 (2012) 170 (http://dx.doi.org/10.1016/j.fitote.2011.10.010)
J. P. Vincken, L. Heng, A. de Grot, H. Gruppen, Phytochem. 68 (2007) 275 (http://dx.doi.org/10.1016/j.phytochem.2006.10.008)
Y. Lu, D. Van, L. Deibert, G. Bishop, J. Balsevich, Phytochem. 113 (2015) 108 (http://dx.doi.org/10.1016/j.phytochem.2014.11.021)
B. Akbari, N. Baghaei‐Yazdi, M. Bahmaie, F. Mahdavi Abhari, BioFactors 48 (2022) 611 (https://doi.org/10.1002/biof.1831)
Q. Z. Lv, J. T. Long, Z. F. Gong, K. Y. Nong, X. M. Liang, T. Qin, W. Huang, L. Yang, Nat. Prod. Commun. 16 (2021) 1 (https://doi.org/10.1177/1934578x211027745)
P. B. Benil, P. Nimisha, S. Arokiyaraj, R. Rajakrishnan, A. Alfarhan, A. Al-Ansari, J. King Saud Univ. Sci. 32 (2020) 2582 (https://doi.org/10.1016/j.jksus.2020.04.016)
A. Lichota, K. Gwozdzinski, Int. J. Mol. Sci. 19 (2018) 3533 (https://doi.org/10.3390/ijms19113533)
F. Morandi, V. Bensa, E. Calarco, F. Pastorino, P. Perri, M. V. Corrias, M. Ponzoni, C. Brignole, Nutrients 13 (2021) 2178 (https://doi.org/10.3390/nu13072178)
A. Kafoud, Z. Salahuddin, R. S. Ibrahim, R. Al-Janahi, A. Mazurakova, P. Kubatka, D. Büsselberg, Biomol. 13 (2023) 563 (https://doi.org/10.3390/biom13030563)
E. Önem, A. G. Özaydın, H. C. Sarısu, Braz. J. Pharm. Sci. 59 (2023) e20412 (http://dx.doi.org/10.1590/s2175-97902023e20412)
S. M. Mandal, R. O. Dias, O. L. Franco, J. Med. Food 20 (2017) 1031 (https://doi.org/10.1089/jmf.2017.0017)
C. Ozay, R. Mammadov, Acta Biol. Hung. 68 (2017) 310 (https://doi.org/10.1556/018.68.2017.3.8)
R. Mammadov, A. Kaska, C. Ozay, Indian J. Pharm. Sci. 79 (2017) 585 (https://doi.org/10.4172/pharmaceutical-sciences.1000266)
C. Ozay, Riv. Ital. Sostanze Gr. 100 (2023) 177 (https://www.innovhub-ssi.it/kdocs/2108136/2023_risg_100_3_4_ozay.pdf)
R. Apak, M. Özyürek, K. Güçlü, E. Çapanoğlu, J. Agric. Food Chem. 64 (2016) 997 (https://doi.org/10.1021/acs.jafc.5b04739)
I. Urkmez, H. I. Karahan Coven, A. Eldem, M. Pehlivan, Eur. J. Biol. 81 (2022) 251 (https://doi.org/10.26650/EurJBiol.2022.1167842)
N. Şirin, L. Elmas, M. Seçme, Y. Dodurga, Gene 737 (2020) 144428 (https://doi.org/10.1016/j.gene.2020.144428)
EUCAST. The European Committee on Antimicrobial Susceptibility Testing, Breakpoint tables for interpretation of MICs and zone diameters, version 13.0, 2023 (http://www.eucast.org)
S. Stepanović, D. Vuković, V. Hola, G. Di Bonaventura, S. Djukić, I. Cirković, F. Ruzicka, APMIS 115 (2007) 891 (https://doi.org/10.1111/j.1600-0463.2007.apm_630.x)
A. Temel, B. Erac, Curr. Microbiol. 79 (2022) 256 (https://doi.org/10.1007/s00284-022-02943-0)
D. B. Singh, R. K. Pathak, D. Rai, Rev. Bras. Farmacogn. 32 (2022) 147 (https://doi.org/10.1007/s43450-022-00235-z)
S. Chandra, D. S. Rawat, Integr. Med. Res. 4 (2015) 123 (https://doi.org/10.1016/j.imr.2015.06.004)
D. Charalambous, M. Christoforou, E. N. Kitiri, M. Andreou, D. Partassides, C. Papachrysostomou, M. Frantzi, G. A. Karikas, M. Pantelidou, Molecules 27 (2022) 5812 (https://doi.org/10.3390/molecules27185812)
N. Kumar, N. Goel, Biotechnol. Rep. 24 (2019) e00370 (https://doi.org/10.1016/j.btre.2019.e00370)
W. Smułek, A. Zdarta, A. Pacholak, A. Zgoła-Grześkowiak, Ł. Marczak, M. Jarzębski, E. Kaczorek, Colloids Surf., B: 150 (2017) 209 (https://doi.org/10.1016/j.colsurfb.2016.11.035)
M. Aijaz, N. Keserwani, M. Yusuf, N.H. Ansari, R. Ushal, P. Kalia, Biointerface Res. Appl. Chem. 13 (2022) 324 (https://doi.org/10.33263/BRIAC134.324)
Z. Qu, A. Liu, P. Li, C. Liu, W. Xiao, J. Huang, Z. Liu, S. Zhang, Crit. Rev. Food Sci. Nutr. 61 (2021) 211 (https://doi.org/10.1080/10408398.2020.1723057)
W. Sun, M. H. Shahrajabian, Molecules 28 (2023) 1845 (https://doi.org/10.3390/molecules28041845)
N. Kimutai, WJPPS 6 (2017) 144 (https://doi.org/10.20959/wjpps20176-9263)
H. Azaizeh, B. Saad, K. Khalil, O. Said, Evid. Based Comp. Alternat. Med. 3 (2006) 229 (https://doi.org/10.1093/ecam/nel034)
J. Kovalevich, D. Langford, in: S.,Amini, M. White, Eds., Neuronal Cell Culture, Methods in Molecular Biology, Humana Press, Totowa, NJ, 2013, pp. 9–21 (https://doi.org/10.1007/978-1-62703-640-5_2)
M. M. Eren, B. Dikmen, C. Vatansever, H. Servi, H. C. Yeğin, G. Ozan, Ann. Med. Res. 28 (2021) 516 (https://dx.doi.org/10.5455/annalsmedres.2020.05.487)
A. Aras, Y. Alan, Erzincan Uni. J Sci. Tech. 15 (2022) 135 (https://doi.org/10.18185/erzifbed.995560).