Quantitative structure-activity relationship modelling of influenza М2 ion channels inhibitors Scientific paper
Main Article Content
Abstract
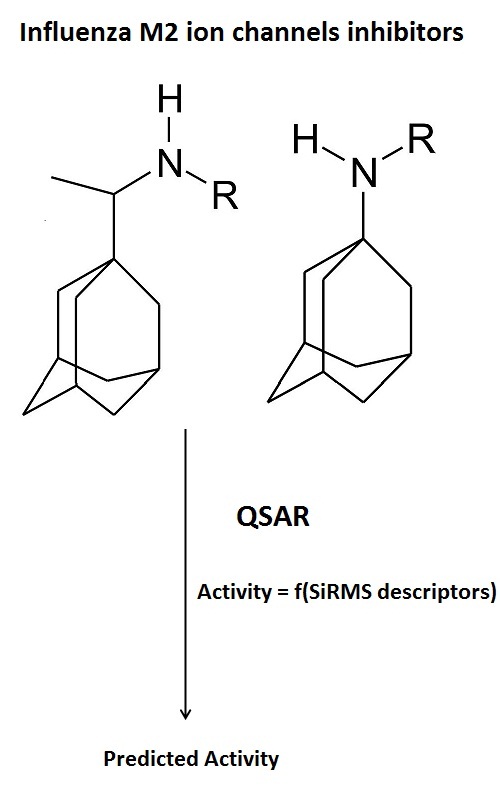
A series of adamantane derivatives (rimantadine and amantadine) incorporating amino-acid residues are investigated by simplex representation of molecular structure (SiRMS) approach in order to found correlation between chemical structures of investigated compounds and obtained data for antiviral activity and cytotoxicity. The obtained data from QSAR analysis show that adamantane derivatives containing amino acids with short aliphatic non-polar residues in the lateral chain will have good antiviral activity against the tested virus A/H3N2, strain Hong Kong/68 with low cytotoxicity. QSAR experiments and in vitro data also show good correlation and reveal that modified adamantine derivatives including guanidated in the lateral chain amino acid and β-amino acids as substituents show low to none activity.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
R. L. Tominack, F. G. Hayden, Infect. Dis. Clin. North. Am. 1 (1987) 459.
R. B. Belshe, M. H. Smith, C. B. Hall, R. Betts, A. J. Hay, J. Virol. 62 (1988) 1508 (https://jvi.asm.org/content/62/5/1508)
J. Wang, C. Ma, J. Wang, H. Jo, B. Canturk, G. Fiorin, W. F. DeGrado, J. Med. Chem. 56 (2013) 2804 (https://doi.org/10.1021/jm301538e)
R. M. Pielak, J. J. Chou, Biochem. Biophys. Res. Commun. 401 (2010) 58 (https://doi.org/10.1016/j.bbrc.2010.09.008)
J. Wang, J. X. Qiu, C. Soto, W. F. DeGrado, Curr. Opin. Struct. Biol. 21 (2011) 68 (https://doi.org/10.1016/j.sbi.2010.12.002)
A. V. Gaiday, I. A. Levandovskiy, K. G. Byler, T. E. Shubina, in Proceedings of International Conference on Computational Science, 2008, Berlin, Germany, pp. 360–368 (https://doi.org/10.1007/978-3-540-69387-1_40)
V. E. Kuzmin, A. G. Artemenko, E. N. Muratov, J. Computer-Aided Molec. Des. 22 (2008) 403 (https://doi.org/10.1007/s10822-008-9211-x)
V. E. Kuzmin, A. G. Artemenko, E. N. Muratov, P. G. Polischuk, L. N. Ognichenko, A. V. Liahovsky, A. I. Hromov, E. V. Varlamova, Recent Advances in QSAR Studies: Methods and Applications,Springer, Dordrecht, 2010, p.127 (ISBN 978-1-4020-9783-6)
S. Rännar, F. Lindgren, P. Geladi, S. Wold, J. Chemometrics 8 (1994) 111 (https://doi.org/10.1002/cem.1180080204)
L. Breiman, Machine Learning 45 (2001) 5 (https://doi.org/10.1023/A:1010933404324)
R. E. Carhart, D. H. Smith, R. Venkataraghavan, J. Chem. Inform. Comp. Sci. 25 (1985) 64 (https://doi.org/10.1021/ci00046a002)
K. Hasegawa, Y. Miyashita, K. Funatsu, J. Chem. Inform. Comp. Sci. 37 (1997) 306 (https://doi.org/10.1021/ci960047x)
V. E. Kuzmin, A. G. Artemenko, P. G. Polischuk, E. N. Muratov, A. I. Hromov, A. V. Liahovskiy, S. A. Andronati, S. Y. Makan, J. Mol. Model 11 (2005) 457 (http://doi.org/10.1007/s00894-005-0237-x)
OECD (2014) Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models, OECD Series on Testing and Assessment, OECD Publishing, Paris, 2014, p.154 (http://doi.org/10.1787/9789264085442-en)
M. Meloun, J. Militku, M. Hill, Analyst 127 (2002) 433 (http://doi.org/10.1039/B110779H)
P. G. Polischuk, E. N. Muratov, A. G. Artemenko, O. G. Kolumbin, N. N. Muratov, V. E. Kuz'min, J. Chem. Inf. Mod. 49 (2009) 2481 (http://doi.org/10.1021/ci900203n)
R. Chayrov, N. A. Parisis, M. V. Chatziathanasiadou, E. Vrontaki, K. Moschovou, G. Melagraki, H. Sbirkova-Dimitrova, B. Shivachev, M. Schmidtke, Y. Mitrev, M. Sticha, T. Mavromoustakos, A. G. Tzakos, I. Stankova, Molecules 25 (2020) 3989 (https://doi.org/10.3390/molecules25173989)
A. Chintakrindi, Ch. D'souza, M. Kanyalkar, Mini Rev. Med. Chem. 12 (2012) 1273 (https://doi.org/10.2174/138955712802761997).