Examination and optimization of lignocellulolytic activity of Stereum gausapatum F28 on beechwood sawdust supplemented with molasses stillage Scientific paper
Main Article Content
Abstract
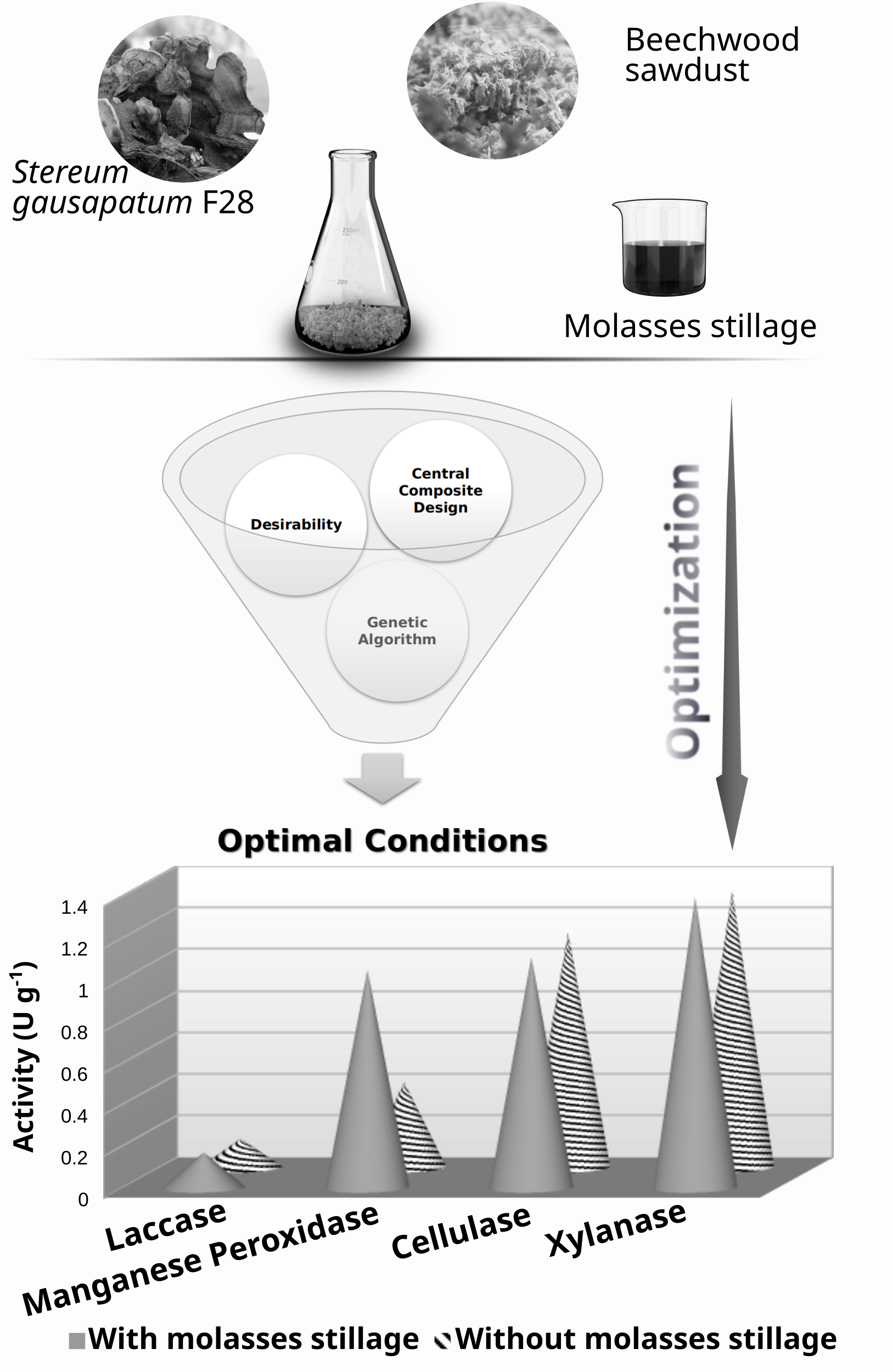
This study provides a detailed analysis of the lignocellulolytic activity of a new isolate Stereum gausapatum F28, a Serbian autochthonous fungi, on beechwood sawdust supplemented with cheap waste, sugar beet molasses stillage. Advanced multiple response optimization techniques were applied to improve ligninolytic and reduce hydrolytic activity as a requirement for potential biorefinery use. The applied techniques were supposed to select cultivation conditions that would give manganese peroxidase and laccase activities above 0.84 and 0.12 U g-1 substrate, respectively, and cellulase and xylanase activities below 1.12 and 1.4 U g-1 substrate. The optimal cultivation conditions that met the set requirements included molasses stillage concentration of 10 %, substrate moisture content of 53 %, incubation temperature of 23.5 °C, and pH 5.2. The research showed that the addition of molasses stillage had a positive effect on enzyme production and that the optimal stillage concentration differed depending on the enzyme type (for laccase it was <5 %, manganese peroxidase ≈12 %, cellulase ≈21 % and xylanase ≈16 %), which should be taken into consideration when optimizing the desired process.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
S. Bharathiraja, J. Suriya, M. Krishnan, P. Manivasagan, S.-K. Kim, in Advances in Food and Nutrition Research, S.-K. Kim, F. Toldrá (Eds.), Academic Press, Cambridge, MA, 2017, pp. 125–148 (https://dx.doi.org/10.1016/bs.afnr.2016.11.003)
A. P. Singh, T. Singh, Biomass Bioenergy 62 (2014) 198 (https://dx.doi.org/10.1016/j.biombioe.2013.12.013)
O. B. Chukwuma, M. Rafatullah, H. A. Tajarudin, N. Ismail, Sustainability 12 (2020) 7282 (https://dx.doi.org/10.3390/su12187282)
W. Mikucka, M. Zielińska, Appl. Biochem. Biotechnol. 192 (2020) 770 (https://dx.doi.org/10.1007/s12010-020-03343-5)
J. Jović, J. Hao, S. Kocić-Tanackov, L. Mojović, Biomass Convers. Biorefinery (2020) (https://dx.doi.org/10.1007/s13399-020-00929-1)
Z. Shahryari, M. H. Fazaelipoor, Y. Ghasemi, P. R. Lennartsson, M. J. Taherzadeh, Molecules 24 (2019) 721 (https://dx.doi.org/10.3390/molecules24040721)
B. K. Acharya, S. Mohana, R. Jog, J. Divecha, D. Madamwar, J. Environ. Manage. 91 (2010) 2019 (https://dx.doi.org/10.1016/j.jenvman.2010.05.001)
V. Singh, S. Haque, R. Niwas, A. Srivastava, M. Pasupuleti, C. K. M. Tripathi, Front. Microbiol. 7 (2017) (https://dx.doi.org/10.3389/fmicb.2016.02087)
D. Palhazi Cuervo, P. Goos, K. Sörensen, Stat. Comput. 26 (2016) 15 (https://dx.doi.org/10.1007/s11222-014-9467-z)
R. Noorossana, S. Davanloo Tajbakhsh, A. Saghaei, Int. J. Adv. Manuf. Technol. 40 (2009) 1227 (https://dx.doi.org/10.1007/s00170-008-1423-7)
J. Jović, A. Buntić, N. Radovanović, B. Petrović, L. Mojović, Food Technol. Biotechnol. 56 (2018) 354 (https://dx.doi.org/10.17113/ftb.56.03.18.5348)
A. Sluiter, B. Hames, D. Hyman, C. Payne, R. Ruiz, C. Scarlata, J. Sluiter, D. Templeton, J. Wolfe, Biomass and Total Dissolved Solids in Liquid Process Samples, Technical Report, National Renewable Energy Laboratory, Golden, CO, 2008, p. 9 (https://www.nrel.gov/docs/gen/fy08/42621.pdf)
R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, 2020 (https://www.R-project.org)
R. V. Lenth, J. Stat. Softw. 32 (2009) 1 (http://dx.doi.org/10.18637/jss.v032.i07)
M. Kuhn, Desirability: function optimization and ranking via desirability functions, R package version 2.1, 2016 (https://CRAN.R-project.org/package=desirability)
L. Scrucca, J. Stat. Softw. 53 (2013) 1 (http://dx.doi.org/10.18637/jss.v053.i04)
L. Boddy, D. W. Bardsley, O. M. Gibbon, New Phytol. 107 (1987) 143 (https://dx.doi.org/https://doi.org/10.1111/j.1469-8137.1987.tb04888.x)
E. Krumova, N. Kostadinova, J. Miteva‐Staleva, G. Stoyancheva, B. Spassova, R. Abrashev, M. Angelova, Eng. Life Sci. 18 (2018) 692 (https://dx.doi.org/https://doi.org/10.1002/elsc.201800055)
J. A. Herrick, Ohio J. Sci. 39 (1939) 254 (https://kb.osu.edu/handle/1811/3038)
C. J. Humphrey, P. V. Siggers, J. Agric. Res. 47 (1933) 997 (https://naldc.nal.usda.gov/download/IND43968250/PDF)
G. C. dos Santos Bazanella, D. F. de Souza, R. Castoldi, R. F. Oliveira, A. Bracht, R. M. Peralta, Folia Microbiol. (Praha) 58 (2013) 641 (https://dx.doi.org/10.1007/s12223-013-0253-7)
P. A. Geethanjali, H. G. Gowtham, M. Jayashankar, Bull. Natl. Res. Cent. 44 (2020) 173 (https://dx.doi.org/10.1186/s42269-020-00426-5)
A. Kumar, Int. J. Microbiol. 2020 (2020) e8894215 (https://dx.doi.org/https://doi.org/10.1155/2020/8894215)
L. M. Legodi, D. La Grange, E. L. J. van Rensburg, I. Ncube, Enzyme Res. 2019 (2019) e1390890 (https://dx.doi.org/https://doi.org/10.1155/2019/1390890)
M. Galbe, O. Wallberg, Biotechnol. Biofuels 12 (2019) 294 (https://dx.doi.org/10.1186/s13068-019-1634-1)
R. Reina, H. Kellner, J. Hess, N. Jehmlich, I. García-Romera, E. Aranda, M. Hofrichter, C. Liers, PLOS ONE 14 (2019) e0212769 (https://dx.doi.org/10.1371/journal.pone.0212769)
A. Tripathi, R. C. Upadhyay, S. Singh, Indian J. Microbiol. 52 (2012) 381 (https://dx.doi.org/10.1007/s12088-011-0232-0)
I. Eichlerová, P. Baldrian, J. Fungi 6 (2020) 301 (https://dx.doi.org/10.3390/jof6040301)
N. Namnuch, A. Thammasittirong, S. N.-R. Thammasittirong, Mycology 12 (2021) 119 (https://dx.doi.org/10.1080/21501203.2020.1806938).