Prediction of retardation factor of protein amino acids in reversed phase TLC with ethanol–sodium azide solution as the mobile phase using QSRR Scientific paper
Main Article Content
Abstract
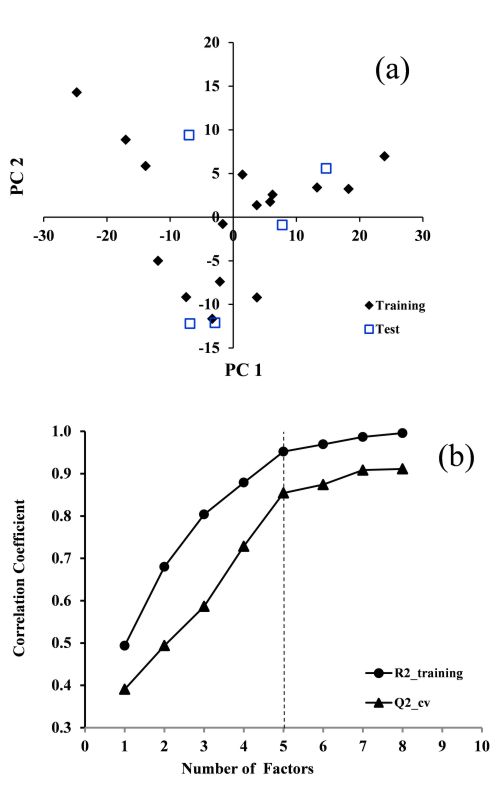
Due to the importance of amino acids (AAs) as the basic bricks of proteins and their application in the drug and food industries, there is great interest in their separation and identification using simple and inexpensive approaches. Application of predictive models for the determination of the behavior of AAs can reduce trial-and-error experiments. Herein, the retardation factor (RF) of 21 protein AAs were studied using the quantitative structure-retardation factor (QSRR) model. The RF values of the AAs in ethanol–sodium azide solution as the mobile phase of reversed phase thin layer chromatography (RP-TLC) were correlated with the structural properties of the AAs. The suggested QSRR indicated excellent fitting and prediction ability (R2train = 0.95 and R2test = 0.94). Furthermore, other statistical tests, such as y-scrambling, cross validation and the Williams plot confirmed the stability, absence of chance and the suitable applicability domain, respectively. It was shown that the sum of geometrical distances between oxygen and nitrogen atoms in an AA molecule is an important factor for the RF values of the AAs in the ethanol–sodium azide.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
S. H. Park, M. De Pra, P. R. Haddad, S. Grosse, C. A. Pohl, F. Steiner, J. Chromatogr., A 1609 (2020) 460508 (https://dx.doi.org/10.1016/j.chroma.2019.460508)
A. M. Ramezani, S. Yousefinejad, A. Shahsavar, A. Mohajeri, G. Absalan, J. Chromatogr., A 1599 (2019) 46 (https://dx.doi.org/10.1016/j.chroma.2019.03.063)
J. M. Sutter, T. A. Peterson, P. C. Jurs, Anal. Chim. Acta 342 (1997) 113 (https://dx.doi.org/10.1016/S0003-2670(96)00578-8)
Y. Marrero-Ponce, S. J. Barigye, M. E. Jorge-Rodríguez, T. Tran-Thi-Thu, Chem. Pap. 72 (2018) 57 (https://dx.doi.org/10.1007/s11696-017-0257-x)
C. Giaginis, A. Tsantili-Kakoulidou, Chromatographia 76 (2013) 211 (https://dx.doi.org/10.1007/s10337-012-2374-6)
J. Dai, L. Jin, S. Yao, L. Wang, Chemosphere 42 (2001) 899 (https://dx.doi.org/10.1016/S0045-6535(00)00181-8)
K. Héberger, J. Chromatogr., A 1158 (2007) 273 (https://dx.doi.org/10.1016/J.CHRO¬MA.2007.03.108)
R. Kaliszan, Chem. Rev. 107 (2007) 3212 (https://dx.doi.org/10.1021/cr068412z)
R. Kaliszan, J. Chromatogr., A 220 (1981) 71 (https://dx.doi.org/10.1016/S0021-9673(00)98504-2)
D. Kaźmierczak, W. Ciesielski, R. Zakrzewski, JPC - J. Planar Chromatogr. - Mod. TLC 18 (2005) 427 (https://dx.doi.org/10.1556/JPC.18.2005.6.5)
T. Hudaib, S. Brown, D. Wilson, P. E. Eady, JPC - J. Planar Chromatogr. - Mod. TLC 29 (2016) 145 (https://dx.doi.org/10.1556/1006.2016.29.2.9)
S. Yousefinejad, F. Honarasa, N. Saeed, J. Sep. Sci. 38 (2015) 1771 (https://dx.doi.org/10.1002/jssc.201401427 )
S. Yousefinejad, F. Honarasa, S. Akbari, M. Nekoeinia, J. Liq. Chromatogr. Relat. Technol. 43 (2020) 580 (https://dx.doi.org/10.1080/10826076.2020.1774388)
R. Todeschini, V. Consonni, Molecular Descriptors for Chemoinformatics, 2nd ed., Wiley-VCH, Weinheim, 2009 (ISBN: 9783527318520)
P. Gramatica, Mol. Inform. 33 (2014) 311 (https://dx.doi.org/10.1002/minf.201400030)
S. Yousefinejad, B. Hemmateenejad, Chemom. Intell. Lab. Syst. 149 (2015) 177 (https://dx.doi.org/10.1016/j.chemolab.2015.06.016)
S. Yousefinejad, F. Honarasa, A. Solhjoo, J. Chem. Eng. Data 61 (2016) 614 (https://dx.doi.org/10.1021/acs.jced.5b00768)
J. U. N. Shao, J. Am. Stat. Assoc. 88 (1993) 486 (https://dx.doi.org/10.2307/2290328)
P. Gemperline, Practical Guide to Chemometrics, 2nd ed., Taylor & Francis Group, Boca Raton, USA, 2006 (ISBN: 1574447831)
F. Honarasa, S. Yousefinejad, S. Nasr, M. Nekoeina, J. Mol. Liq. 212 (2015) 52 (https://dx.doi.org/10.1016/j.molliq.2015.08.055)
A. Golbraikh, A. Tropsha, Mol. Divers. 5 (2000) 231 (https://dx.doi.org/10.1023/A:1021372108686)
K. Roy, R. N. Das, P. Ambure, R. B. Aher, Chemom. Intell. Lab. Syst. 152 (2016) 18 (https://dx.doi.org/10.1016/j.chemolab.2016.01.008)
K. Roy, P. Chakraborty, I. Mitra, P. K. Ojha, S. Kar, R. N. Das, J. Comput. Chem. 34 (2013) 1071 (https://dx.doi.org/10.1002/jcc.23231)
L. Eriksson, J. Jaworska, A. P. Worth, M. T. D. Cronin, R. M. McDowell, P. Gramatica, Environ. Health Perspect. 111 (2003) 1361 (https://dx.doi.org/10.1289/ehp.5758)
T. A. Craney, J. G. Surles, Qual. Eng. 14 (2002) 391 (https://dx.doi.org/10.1081/QEN-120001878)
R. Todeschini, V. Consonni, P. Gramatica, in Comprehensive Chemometrics: Chemical and Biochemical Data Analysis, R. Tauler, B. Walczak, S. D. Brown (Eds.), Elsevier B.V., Amsterdam, 2009, pp. 129–172 (ISBN: 9780444641663)
T. I. Netzeva, A. P. Worth, T. Aldenberg, R. Benigni, M. T. D. Cronin, P. Gramatica, J. S. Jaworska, S. Kahn, G. Klopman, C. A. Marchant, G. Myatt, N. Nikolova-Jeliazkova, G. Y. Patlewicz, R. Perkins, D. W. Roberts, T. W. Schultz, D. T. Stanton, J. J. M. van de Sandt, W. Tong, G. Veith, C. Yang, Altern. to Lab. Anim. 33 (2005) 155 (https://dx.doi.org/10.1177/026119290503300209)
S. Yousefinejad, R. Eftekhari, F. Honarasa, Z. Zamanian, F. Sedaghati, J. Mol. Liq. 241 (2017) 861 (https://dx.doi.org/10.1016/j.molliq.2017.06.081).