Impaired local hydrophobicity, structural stability and conformational flexibility due to point mutations in SULT1 family of enzymes Scientific paper
Main Article Content
Abstract
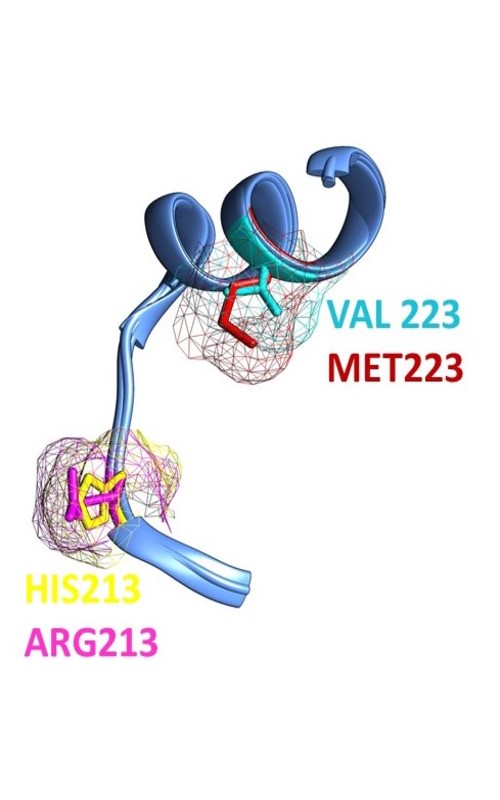
Sulfotransferases (SULTs) are enzymes involved in phase II of the metabolism of xenobiotics. Single nucleotide polymorphisms were identified for genes encoding the SULTs leading to allozymes with modified sulfating activity. This study aims to analyse the effects of the most frequently identified amino acid mutations in the sequences of enzymes belonging to the SULT1 family on their local properties and structural stability. The outcomes reveal that single point mutations alter the local hydrophobicity and flexibility, mainly due to destabilization of the protein structures, may consequently lead to changes in the dynamic of the active site activity reducing the affinity for the substrate. Elucidation of how the single point mutations influence the activity of enzymes contributes to understanding the molecular basis of the specificity of enzymatic activity and mitigating anomalies in the metabolism of xenobiotics.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
M. W. Duffel, Comprehensive Toxicology, Elsevier, Amsterdam, 2010, p. 367 (https://doi.org/10.1016/B978-0-08-046884-6.00418-8)
H. Glatt, W. Meinl, Naunyn-Schmiedeb, Arch. Pharmacol. 55 (2004) 369 (http://doi.org/10.1007/s00210-003-0826-0)
M.W.H. Coughtrie, Chem. Biol. Interact. 259 (2016) 2 (http://doi.org/10.1016/j.cbi.2016.05.005)
Z. Riches, E. L. Stanley, J. C. Bloomer & M. W. H. Coughtrie, Drug Metab. Dispos. 37 (2009) 2255 (http://doi.org/10.1124/dmd.109.028399)
Z. Riches, J.C. Bloomer, M.W.H. Coughtrie, Biochem. Pharmacol. 74 (2007) 352 (http://doi.org/10.1016/j.bcp.2007.04.006)
A. F. Bairam, M. I. Rasool, F. A. Alherz, M. S. Abunnaja, A. A. El Daibani, S. A. Gohal, M.-C. Liu, Biochem. Pharmacol. 151 (2018) 104 (http://doi.org/10.1016/j.bcp.2018.03.005)
A.C.S. Barbosa, Y. Feng, C. Yu, M. Huang, W. Xie, Expert. Opin. Drug. Metab. Toxicol. 15 (2019) 329 (http://doi.org/10.1080/17425255.2019.1588884)
K. Kurogi, M. I. Rasool, F. A. Alherz, A. A. El Daibani, A. F. Bairam, M. S. Abunnaja, M.-C. Liu, Expert. Opin. Drug. Metab. Toxicol. 17 (2021) 767 (http://doi.org/10.1080/17425255.2021.1940952)
K.-A. Kim, S.-Y. Lee, P.-W. Park, J.-M. Ha, J.-Y. Park, Eur. J. Clin. Pharmacol. 61 (2005) 743 (http://doi.org/10.1007/s00228-005-0989-3)
N. Hempel, N. Gamage, J. L. Martin & M. E. McManus, Int. J. Biochem. Cell Biol. 39 (2007) 685 (http://doi.org/10.1016/j.biocel.2006.10.002)
S.-J. Lee, W.-Y. Kim, Y. B. Jarrar, Y.-W. Kim, S. S. Lee, J.-G. Shin, Drug Metab. Pharmacokinet. 28 (2013) 372 (http://doi.org/10.2133/dmpk.dmpk-12-sc-110)
S. Nagar, S. Walther, R. L. Blanchard, Mol. Pharmacol. 69 2006 2084 (http://doi.org/10.1124/mol.105.019240)
M.I. Rasool, A.F. Bairam, S.A. Gohal, A.A. El Daibani, F.A. Alherz, M.S. Abunnaja, E.S. Alatwi, K. Kurogi, M.C. Liu, Pharmacol. Rep. 71 (2019) 257 (http://doi.org/10.1016/j.pharep.2018.12.001)
W. Meinl, J. H. Meerman, H. Glatt, Pharmacogenetics 12 (2002) 677 (http://doi.org/10.1097/00008571-200212000-00002)
Y. Hui, M.-C. Liu, Eur. J. Pharmacol. 761 (2015) 125 (http://doi.org/10.1016/j.ejphar.2015.04.039)
A. F. Bairam, M. I. Rasool, F. A. Alherz, M. S. Abunnaja, A. A. El Daibani, K. Kurogi & M.-C. Liu, Arch. Biochem. Biophys. 648 (2018b) 44 (http://doi.org/10.1016/j.abb.2018.04.019)
Z. E. Tibbs, A. L. Guidry, J. L. Falany, S. A. Kadlubar, C. N. Falany, Xenobiotica 48 (2017) 79 (http://doi.org/10.1080/00498254.2017.1282646)
R. R. Freimuth, B. Eckloff, E. D. Wieben, R. M. Weinshilboum, Pharmacogenetics 11 (2001) 747 (http://doi.org/10.1097/00008571-200112000-00002)
A. A. Adjei, B. A. Thomae, J. L. Prondzinski, B. W. Eckloff, E. D. Wieben, R. M. Weinshilboum, Br. J. Pharmacol. 139 (2003) 1373 (http://doi.org/10.1038/sj.bjp.0705369)
A. A. El Daibani, F. A. Alherz, M. S. Abunnaja, A. F. Bairam, M. I. Rasool, K. Kurogi, M.-C. Liu, Eur. J. Drug Metab. Pharmacokinet. 46 (2020) 105 (http://doi.org/10.1007/s13318-020-00653-1)
A. Isvoran, & Y. Peng, S. Ceauranu, L. Schmidt, A. Nicot, M. Miteva, Drug Discov. Today. 27 (2022) 103349 (http://doi.org/10.1016/j.drudis.2022.103349)
L. M. Bidwell, M. E. McManus, A. Gaedigk, Y. Kakuta, M. Negishi, L. Pedersen, J. L. Martin, J. Mol. Biol. 293 (1999) 521 (http://doi.org/10.1006/jmbi.1999.3153)
N. U. Gamage, S. Tsvetanov, R. G. Duggleby, M. E. McManus and J. L. Martin, J. Biol. Chem. 280 (2005) 41482 (http://doi.org/10.1074/jbc.m508289200)
J.-H. Lu, H.-T. Li, M.-C. Liu, J.-P. Zhang, M. Li, X.-M. An & W.-R. Chang, Biochem. Biophys. Res. Commun. 335 (2005) 417 (http://doi.org/10.1016/j.bbrc.2005.07.091)
J. Lu, H. Li, J. Zhang, M. Li, M.-Y. Liu, X. An, W. Chang, Biochem. Biophys. Res. Commun. 396 (2010) 429 (http://doi.org/10.1016/j.bbrc.2010.04.109)
I. Berger, C. Guttman, D. Amar, R. Zarivach, A. Aharoni, PLoS ONE 6 (2011) e26794 (http://doi.org/10.1371/journal.pone.0026794)
R. A. Gosavi, G. A. Knudsen, L. S. Birnbaum, L. C. Pedersen, Environ. Health Perspect. (2013) (http://doi.org/10.1289/ehp.1306902)
Z. E. Tibbs, C. N. Falany, Pharmacol. Res. Perspect. 3 (2015) e00147 (http://doi.org/10.1002/prp2.147)
K. Evgeny, Bioinformatics 23 (2007) 717 (http://doi.org/10.1093/bioinformatics/btm006)
C. H. M. Rodrigues, D. E. V. Pires, D. B. Ascher, Protein Sci. (2020) (http://doi.org/10.1002/pro.3942)
A. Bateman, M.-J. Martin, S. Orchard, M. Magrane, R. Agivetova, S. Ahmad, E. Alpi, E. H. Bowler-Barnett, R. Britto, B. Bursteinas, H. Bye-A-Jee, R. Coetzee, A. Cukura, A. Da Silva, P. Denny, T. Dogan, T. Ebenezer, J. Fan, D.Teodoro, Nucleic Acids Res. 49 (2020) 480 (http://doi.org/10.1093/nar/gkaa1100)
M. R. Wilkins, E. Gasteiger, A. Bairoch, J. C. Sanchez, K. L. Williams, R. D. Appel, D. F. Hochstrasser, Methods Mol Biol. 112 (1999) 531 (http://doi.org/10.1385/1-59259-584-7:531)
T. Hrabe, Z. Li, M. Sedova, P. Rotkiewicz, L. Jaroszewski, A.Godzik, Nucleic Acids Res. 44 (2015) 423 (http://doi.org/10.1093/nar/gkv1316)
H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, Nucleic Acids Res. 28 (2000) 235 (http://doi.org/10.1093/nar/28.1.235)
E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, J. Comput. Chem. 25 (2004) 1605 (http://doi.org/10.1002/jcc.20084)
I. Cook, T. Wang, S. C. Almo, J. Kim, C. N. Falany, T.s S. Leyh, Biochemistry 52 (2013) 415 (https://doi.org/10.1021/bi301492j)
N. U. Gamage, R. G. Duggleby, A. C. Barnett, M. Tresillian, C. F. Latham, N. E. Liyou, M. E. McManus, J. L. Martin, J. Biol. Chem. 278 (2003) 7655. (https://doi.org/10.1074/jbc.M207246200)
U. Alcolombri, M. Elias, D. S. Tawfik, J. Mol. Biol. 411 (2011) 837 (https://doi.org/10.1016/j.jmb.2011.06.037)
I. Cook, T. Wang, T. S. Leyh, Biochemistry 54 (2015) 6114 (https://doi.org/10.1021/acs.biochem.5b00406)
R. Dash, M. C. Ali, N. Dash, M. A. K. Azad, S. M. Z. Hosen, M. A. Hannan, I. S. Moon, Int. J. Mol. Sci. 20 (2019) 6256 (http://doi.org/10.3390/ijms20246256)
S. Zhao, D. S. Goodsell, A. J. Olson, Proteins: Struct. Funct. Gen. 43 (2001) 271 (http://doi.org/10.1002/prot.1038)
D. Craciun, A. Isvoran N. M. Avram, Rom. J. Phys. 56 (2011) 185 (https://rjp.nipne.ro/2011_56_1-2/0185_0195.pdf)
D. Craciun, A. Isvoran N. M. Avram, AIP Conf. Proc. 1262 (2010) 173 (http://doi.org/10.1063/1.3482227)
D. Craciun, A. Isvoran N. M. Avram, Phys., A 388 (2009) 4609 (http://doi.org/10.1016/j.physa.2009.07.042)
A. Ciorsac, D. Craciun, V. Ostafe, A. Isvoran, Chaos Solit. Fractals 44 (2011) 191 (http://doi.org/10.1016/j.chaos.2011.01.008)
Y. Zhu, S. Fu, C. Wu, B. Qi, F. Teng, Z. Wang, L. Jiang, Food Hydrocoll. (2020) 105709 (http://doi.org/10.1016/j.foodhyd.2020.105709)
N. Hempel, M. Negishi, M. E. McManus, Methods Enzymol. 400 (2005) 147 (http://doi.org/10.1016/s0076-6879(05)00009-1)
M. Y. Lobanov, E. I. Furletova, N. S. Bogatyreva, M. A. Roytberg, O. V. Galzitskaya, PLoS Comput. Biol. 6 (2010) e1000958 (http://doi.org/10.1371/journal.pcbi.1000958)
N. Sinha, S. Smith-Gill, Curr. Protein Pept. Sci. 3 (2002) 601 (http://doi.org/10.2174/1389203023380431)
A. Isvoran, C. T. Craescu, E. Alexov, Eur. Biophys. J. 36 (2007) 225 (http://doi.org/10.1007/s00249-006-0123-1)
R. L. Redler, J. Das, J. R. Diaz, N. V. Dokholyan, J. Mol. Evol. 82 (2015) 11 (http://doi.org/10.1007/s00239-015-9717-5)
D. M. Popović, I. S. Đorđević, J. Serb. Chem. Soc. 85 (2020) 1429 (https://doi.org/10.2298/JSC200720047P)
D. V. Makhov, D. M. Popović, A. A. Stuchebrukhov, J. Phys. Chem., B 110 (2006) 12162 (https://doi.org/10.1021/jp0608630)
A. Isvoran, M. Louet, D. L. Vladoiu, D. Craciun, M.-A. Loriot, B. O. Villoutreix, M. A. Miteva, Drug Discov. Today 22 (2017) 366 (http://doi.org/10.1016/j.drudis.2016.09.015).