Exploring the efficacy of natural compounds against SARS-CoV-2: A synergistic approach integrating molecular docking and dynamic simulation Scientific paper
Main Article Content
Abstract
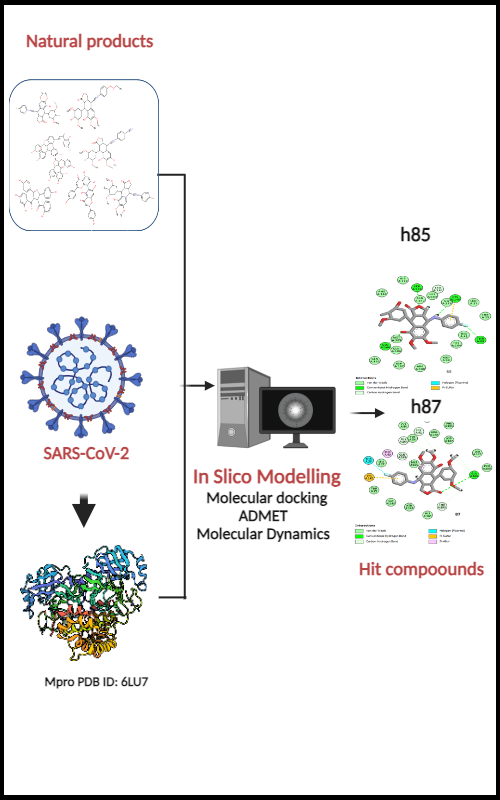
The primary aim of the current investigation is to contribute to SARS-CoV-2 research by identifying potential lead compounds for clinical applications, with a specific focus on inhibitors targeting the main protease (Mpro). In this research, molecular docking analysis was conducted using the software molecular operating environmental (MOE) to evaluate the potency of bioactive compounds sourced from medicinal plants as inhibitors of SARS-CoV-2 Mpro. Among 118 natural compounds with anti-HIV characteristics, the top seven candidates (h3, h84, h85, h87, h90, h108 and h110), were identified based on their superior binding energies with comparison to the reference ligand N3. These selected compounds exhibited binding affinities of –33.996, –35.336, –32.615, –32.154, –33.452, –31.903 and –40.360 kJ mol-1, respectively. To further refine our shortlist of potential candidates for human application, we examined the drug-likeness, and the pharmaceutical attributes of these compounds using the SwissADME web server. Among them, only two compounds, namely h85 and h87, demonstrated favorable pharmacological properties suitable for human administration. These two compounds were subsequently shortlisted for further investigation. To explore the conformational stability of ligands within the Mpro active site, we performed molecular dynamics (MD) simulations. These simulations showed reliable and steady trajectories, supported by analyses of root-mean-square-fluctuation (RMSF) and root-mean-square deviation (RMSD). These findings and favorable molecular properties as well as interaction profiles suggest that these two lead compounds may be promising SARS-CoV-2 therapeutic candidates. They present exciting starting points for further drug design.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou, J. He, L. Liu, H. Shan, C. Lei, D. Hui, B. Du, L. Li, G. Zeng, K. Yuen, R. Chen, C. Tang, T. Wang, P. Chen, J. Xiang, S. Li, J. L. Wang, Z. Liang, Y. Peng, L. Wei, Y. Liu, Y. H. Hu, P. Peng, J. M. Wang, J. Liu, Z. Chen, G. Li, Z .Zheng, S. Qiu, J. Luo, C. Ye , S. Zhu, N. Zhong, N. Eng. J. Med. 382 (2020) 17085 (https://doi.org/10.1056/NEJMoa2002032)
C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, B. Cao, Lancet 395 (2020) 497 (https://doi.org/10.1016/S0140-6736(20)30183-5)
Y. C. Wu, C. S. Chen,Y. J. Chan. J. Chinese Med. Assoc. 83 (2020) 217 (https://doi.org/10.1097/JCMA.0000000000000270)
Z.Y.Zu, M. D. Jiang, P. P. Xu, W. Chen, Q. Q. Ni, G. M. Lu, L. J. Zhang, Radiology 296 (2020) E15 (https://doi.org/10.1148/radiol.2020200490 )
A. A. T. Naqvi, K. Fatima, T. Mohammad, U. Fatima, I. K. Singh, A. Singh, S. M . Atif, G. Hariprasad, G. M. Hasan, M. I. Hassan, Mol. Basis Dis. 1866 (2020) 165878 (https://doi.org/10.1016/j.bbadis.2020.165878 )
S.Mahmud, S.Biswas, G. Kumar Paul, A. M. Mita, S. Afrose, M. Robiul Hasan, M. Sharmin Sultana Shimu, M. A. R. Uddin, M. Salah Uddin, S. Zaman, K. M. Kaderi Kibria, M. Arif Khan, T. Bin Emran, M. Abu Saleh, Arab. J. Chem. 14 (2021) 103315 ( https://doi.org/10.1016/j.arabjc.2021.103315)
N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F Gao, W.A. Tan, N. Eng. J. Med. 382 (2020) 727 (https://doi.org/10.1056/NEJMoa2001017)
X. Cui, Y. Wang, J. Zhai, M. Xue, C. Zheng, L. Yu, Virus Research 328 (2023) 199075 (https://doi.org/10.1016/j.virusres.2023.199075)
R. Gili, R. Burioni, J. Transl. Med. 21 (2023) 251(https://doi.org/10.1186/s12967-023-04095-6)
WHO world Health Organization [WHO], Tracking SARS-CoV-2 variants, 2023, https://www. who.int/activities/tracking-SARS-CoV-2-variants
W. T. Harvey, A. M. Carabelli, B. Jackson, R. K. Gupta, E. C. Thomson, E. M. Harrison, C. A. Ludden, R. Reeve, A. Rambaut, COVID-19 Genomics UK (COG-UK) Consortium, S. J. Peacock, D. L. Robertson, Nat. Rev. Microbiol. 19 (2021) 409 (https://doi.org/10.1038/s41579-021-00573-0)
S. O. Aftab, M. Z. Ghouri, M. U. Masood, Z. Haider, Z. Khan, A. Ahmad, N. Munawar, Transl. Med. 18 (2020) 275(https://doi.org/10.1186/s12967-020-02439-0)
R. Yu, L. Chen, R. Lan, R. Shen, P. Li, Int. J. Antimicrob. Agents 56 (2020) 106012 (https://doi.org/10.1016/j.ijantimicag.2020.106012)
M. Lounasmaa, P. Hanhunen, M. Westersund, N. Halonen, Alkaloids: Chem. Biol. 52 (1999) 103(https://doi.org/10.1016/S0099-9598(08)60026-7)
R. M. Perez, Pharm. Biol. 41 (2003) 107 (https://doi.org/10.1076/phbi.41.2.107.14240)
S. Chtita, R. T. Fouedjou, S. Belaidi, L. A. Djoumbissie, M. Ouassaf, F. A. Qais, M Bakhouch, M. Efendi, T. T. Tok, M. Bouachrine, T. Lakhlifi, Struct Chem. 33 (2022) 1799 (https://doi.org/10.1007/s11224-022-01939-7)
J. G. Africa, H. C. Arturo, L. J. Bernardo, J. K. Ching, O. C. de la Cruz, J. B. Hernandez, R. J. Magsipoc, C. T. Sales, J. C. Agbay, G. L. Neri, M. T. Quimque, A. P. Macabeo, Philipp. J. Sci. 151 (2021) 35 (https://doi.org/10.56899/151.01.04)
P. Gale, Micro. Risk. Anal. 21 (2022)100198 (https://doi.org/10.1016/j.mran.2021.100198)
P. Gale, Micro. Risk. Anal. 16 (2020) 100140 (https://doi.org/10.1016/j.mran.2020.100140)
M. Popovic, Micro. Risk. Anal. 23 (2023) 100250 (https://doi.org/10.1016/j.mran.2023.100250)
M. Popovic and M. Popovic, Micro. Risk. Anal. 21 (2022)100202 (https://doi.org/10.1016/j.mran.2022.100202)
M. Popovic, Micro.Risk.Anal.24 (2023) 100260 (https://doi.org/10.1016/j.mran.2023.100260)
M. E. Popovic, M. P. Pavlovic, M. Papovic, Micro. Risk. Anal. 25 (2023) 100280
(https://doi.org/10.1016/j.mran.2023.100280)
B. Hemmateenejad, K. Javidnia, M. Nematollahi, M. Elyasi, J. Iran. Chem. Soc. 6 (2009) 420 (https://doi.org/10.1007/BF03245853)
A. Aouidate, A. Ghaleb, S. Chtita, M. Aarjane, A. Ousaa, H. Maghat, A. Sbai, M. Choukrad, M. Bouachrine, T. Lakhlifi, J. Biomol. Struct. Dyn. 39 (2021)4522 (https://doi.org/10.1080/07391102.2020.1779130)
R. Banerjee, L. Perera, L. M. V. Tillekeratne, Drug .Discov. Today 26 (2021) 804 (https://doi.org/10.1016/j.drudis.2020.12.005)
P. K. Doharey, V. Singh, M. R. Gedda, A. K. Sahoo, P. K. Varadwaj, B. Sharma, J. Biomol. Struct. Dyn. 40 (2022) 5588 (https://doi.org/10.1080/07391102.2021.1871956)
M. T. J. Quimque, K. I. R. Notarte, R. A. T. Fernandez, M. A. O. Mendoza, R. A. D. Liman, J. A. K. Lim, L. A. E. Pilapil, J. K. H. Ong, A. M. Pastrana, A. Khan, D. Q. Wei, A. P. G. Macabeo, J. Biomol. Struct. Dyn. 39 (2021) 4316 (https://doi.org/10.1080/07391102.2020.1776639)
V. N. O. de Leon, J. A. H. Manzano, D. Y. H. Pilapil, R. A. T. Fernandez, J. K. A. R. Ching, M. T. J. Quimque, J.C.M. Agbay, K. I. R. Notarte, A. P. G. Macabeo, J. Genet. Eng. Biotechnol. 19 (2021) 104 (https://doi.org/10.1186/s43141-021-00206-2)
D. Li, J. Luan, L. Zhang, Biochem. Biophys. Res. Commun. 538 (2021) 72 (https://doi.org/10.1016/j.bbrc.2020.11.083)
HYPERCHEM Molecular Modeling System, Hypercube. Inc., Gainesville, FL, 2007
MARVINSKETCH 17.1.2, ChemAxon 2017 (http://www.chemaxon.com)
S. Belaidi, R. Mazri, H. Belaidi, T. Lanez, D. Bouzidi, Asian J. Chem. 25 (2013) 9241 (https://doi.org/10.14233/ajchem.2013.15199)
A. Kerassa, S. Belaidi, D. Harkati, T. Lanez, O. Prasad, L. Sinha, Rev. Theor. Sci. 4 (2016) 85 (https://doi.org/10.1166/rits.2016.1050)
S. Chtita, A.Belhassan, A. Aouidate, S. Belaidi, M. Bouachrine, T. Lakhlifi, Comb. Chem. High Throughput Screen. 24 (2021) 441 (https://doi.org/10.2174/1386207323999200730205447)
Molecular Operating Environment (MOE), Version 2007.09, Chemical Computing Group, Inc., Montreal, Quebec, 2005 (http://www.Chemcomp.com)
P. S. Das, A. Kokardekar, C. M. Breneman, J. Chem. Inf. Model. 49 (2009) 2863 (https://doi.org/10.1021/ci900317x)
M. Ouassaf, S. Belaidi, S. Khamouli, H. Belaid, S. Chtita, Acta Chim. Slov. 68 (2021) 289 (https://doi.org/10.17344/acsi.2020.5985)
C. A. Lipinski, F. Lombardo, B. W. Dominy, P. J. Feeney, Adv. Drug. Deliv. Rev. 46 (2001) 3 (https://doi.org/10.1016/s0169-409x(00)00129-0)
A. Daina, O. Michielin, V. Zoete, Sci. Rep. 7 (2017) 42717 (https://doi.org/10.1038/srep42717)
Schrödinger Release 2021-3: Maestro-Desmond Interoperability Tools, Schrödinger, LLC, New York
K. Roos, C. Wu, W. Damm, M. RebouL, J. M. Stevenson, C. Lu, M. K. Dahlgren, S. Mondal, W. Chen, L. Wang, R. Abel, R. A. Friesner, E. D. Harder, J. Chem. Theory Comput. 15 (2019) 1863(https://doi.org/10.1021/acs.jctc.8b01026)
A. Imberty, C. Gautier, J. Lescar, S. Pérez, L. Wyns, R. Loris, J. Biol. Chem. 275 (2000) 17541 (https://doi.org/10.1074/jbc.M000560200)
K. O. Chang, Y. Kim, S. Lovell, A. D. Rathnayake, W. C. Groutas, Viruses 11 (2019) 197 (https://doi.org/10.3390/v11020197)
D. F. Veber, S. R. Johnson, H. Y. Cheng, B. R. Smith, K. W. Ward, K. D. Kopple, J. Med. Chem. 45 (2002) 2615 (https://doi.org/10.1021/jm020017n)
A. K. Ghose, V. N. Viswanadhan, J. J. Wendoloski, J. Comb. Chem. 1 (1999) 55 (https://doi.org/10.1021/cc9800071)
I. Muegge, S. L. Heald, D. Brittelli, J. Med. Chem. 44 (2001) 1841 (https://doi.org/10.1021/jm015507e)
W. J. Egan, K. M. Merz, J. J. Baldwin, J. Med. Chem. 43 (2000) 3867 (https://doi.org/10.1021/jm000292e)
A. Zerroug , S. Belaidi, I. BenBrahim, L. Sinha, S. Chtita, J. King Saud Univ. Sci. 31 (2019) 595-560 (https://doi.org/10.1016/j.jksus.2018.03.024)
N. Aoumeur, S. Belaidi, N. Tchouar, M. Ouassaf, T. Lanez, S. Chtita, Mor. J. Chem. 9 (2021) 274 (https://doi.org/10.48317/IMIST.PRSM/morjchem-v9i2.19884)
H. Nour, O. Daoui, O. Abchir, S. ElKhattabi, S. Belaidi, S. Chtita, Heliyon 8 (2022) e11991 (https://doi.org/10.1016/j.heliyon.2022.e11991)
A. Daina, V. Zoete, J. Med. Chem. 6 (2016) 1117 (https://doi.org/10.1002/cmdc.201600182)
V. Zoete, A. Daina, C. Bovigny, O. Michielin, J. Chem. Info. Model. 56 (2016) 1399 (https://doi.org/10.1021/acs.jcim.6b00174)
S. Ghahremanian, M. M. Rashidi, K. Raeisi, D. Toghraie. J. Mol. Liq. 354 (2022) 118901 (https://doi.org/10.1016/j.molliq.2022.118901).