Identification of musk compounds as inhibitors of the main SARS-CoV-2 protease by molecular docking and molecular dynamics studies Scientific paper
Main Article Content
Abstract
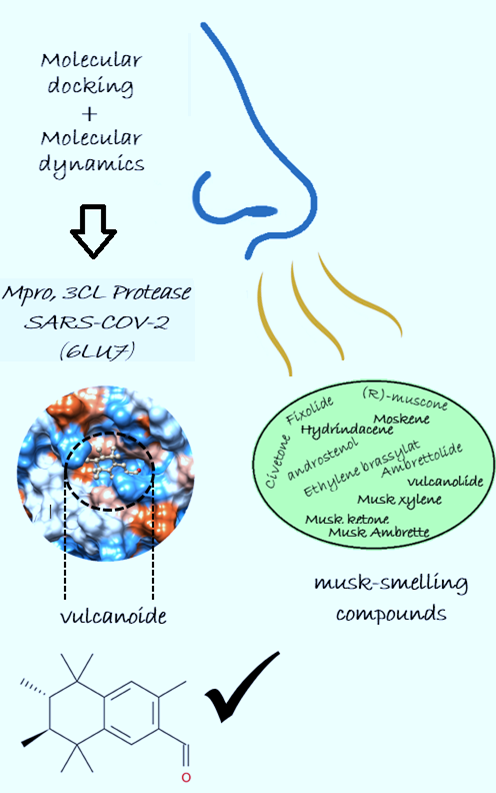
As new drug development is a long process, reuse of bioactives may be the answer to new epidemics; thus, screening existing bioactive compounds against a new SARS-CoV-2 infection is an important task. With this in mind, we have systematically screened potential odorant molecules in the treatment of this infection based on the affinity of the selected odorant compounds on the studied enzyme and the sequence identity of their target proteins (olfactory receptors) to the same enzyme (the main protease of SARS-CoV-2). A total of 12 musk odorant compounds were subjected to a molecular docking and molecular dynamics study to predict their impact against the main protease of SARS-CoV-2. In this study, we have identified two musk-scented compounds (androstenol and vulcanolide) that have good binding energy at the major protease binding site of SARS-CoV-2. However, the RMSD values recorded during dynamic simulation show that vulcanolide exhibits high stability of the protein–ligand complex compared to androstenol. The perspectives of this work are as follows: in vitro, in vivo and clinical trials to verify the computational findings.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Universidad Católica de la Santísima Concepción
Grant numbers DIREG 03/2020 -
Consejo Nacional de Ciencia y Tecnología
Grant numbers 202203072N
References
R. Tosepu, J. Gunawan, D. S. Effendy, L. O. A. I. Ahmad, H. Lestari, H. Bahar, P. Asfian, Sci. Total Environ. 725 (2020) 138436 (http://dx.doi.org/10.1016/J.SCITOTENV.2020.138436)
C. Wu, Y. Liu, Y. Yang, P. Zhang, W. Zhong, Y. Wang, Q. Wang, Y. Xu, M. Li, X. Li, M. Zheng, L. Chen, H. Li, Acta Pharm. Sin., B 10 (2020) 766 (http://dx.doi.org/10.1016/J.APSB.2020.02.008)
A. K. Singh, A. Singh, A. Shaikh, R. Singh, A. Misra, Diabetes Metab. Syndr. Clin. Res. Rev. 14 (2020) 241 (http://dx.doi.org/10.1016/J.DSX.2020.03.011)
D. Kang, H. Choi, J. H. Kim, J. Choi, Int. J. Infect. Dis. 94 (2020) 96 (http://dx.doi.org/10.1016/j.ijid.2020.03.076)
P. Zhai, Y. Ding, X. Wu, J. Long, Y. Zhong, Y. Li, Int. J. Antimicrob. Agents 55 (2020) 105955 (http://dx.doi.org/10.1016/J.IJANTIMICAG.2020.105955)
Y. Shi, J. Wang, Y. Yang, Z. Wang, G. Wang, K. Hashimoto, K. Zhang, H. Liu, Brain, Behav. Immun. – Heal. 4 (2020) 100064 (http://dx.doi.org/10.1016/J.BBIH.2020.100064)
C. H. Parga-Lozano, Biomed. J. Sci. Tech. Res. 35 (2021) 28000 (http://dx.doi.org/10.26717/bjstr.2021.35.005761)
Z. Wang, X. Chen, Y. Lu, F. Chen, W. Zhang, Biosci. Trends 14 (2020) 64 (http://dx.doi.org/10.5582/BST.2020.01030)
A. A. Elfiky, Life Sci. 248 (2020) 117477 (https://doi.org/10.1016/J.LFS.2020.117477)
B. Robson, Comput. Biol. Med. 119 (2020) 103670 (http://dx.doi.org/10.1016/J.COMPBIOMED.2020.103670)
V. Pooladanda, S. Thatikonda, C. Godugu, Life Sci. 254 (2020) 117765 (http://dx.doi.org/10.1016/J.LFS.2020.117765)
R. Hatada, K. Okuwaki, Y. Mochizuki, K. Fukuzawa, Y. Komeiji, Y. Okiyama, S. Tanaka, J. Chem. Inf. Model. 60 (2020) 3593 (https://doi.org/10.1021/acs.jcim.0c00283)
X. Tang, R. H. Du, R. Wang, T. Z. Cao, L. L. Guan, C. Q. Yang, Q. Zhu, M. Hu, X. Y. Li, Y. Li, L. R. Liang, Z. H. Tong, B. Sun, P. Peng, H. Z. Shi, Chest 158 (2020) 195 (http://dx.doi.org/10.1016/j.chest.2020.03.032)
J. Fantini, C. Di Scala, H. Chahinian, N. Yahi, Int. J. Antimicrob. Agents 55 (2020) 105960 (http://dx.doi.org/10.1016/J.IJANTIMICAG.2020.105960)
Z. Sahraei, M. Shabani, S. Shokouhi, A. Saffaei, Int. J. Antimicrob. Agents 55 (2020) 105945 (http://dx.doi.org/10.1016/J.IJANTIMICAG.2020.105945)
A. C. Tsang, S. Ahmadi, J. Hamilton, J. Gao, G. Virgili, S. G. Coupland, C. C. Gottlieb, Am. J. Ophthalmol. 206 (2019) 132 (http://dx.doi.org/10.1016/j.ajo.2019.04.025)
P. Gautret, J. C. Lagier, P. Parola, V. T. Hoang, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V. E. Vieira, H. Tissot Dupont, S. Honoré, P. Colson, E. Chabrière, B. La Scola, J.-M. Rolain, P. Brouqui, D. Raoult, Int. J. Antimicrob. Agents 56 (2020) 105949 (http://dx.doi.org/10.1016/J.IJANTIMICAG.2020.105949)
J. B. Radke, J. M. Kingery, J. Maakestad, M. D. Krasowski, Toxicol. Reports 6 (2019) 1040 (http://dx.doi.org/10.1016/J.TOXREP.2019.10.006)
C. Colalto, Drug Dev. Res. 81 (2020) 950 (http://dx.doi.org/10.1002/ddr.21716)
E. O. Ojah, Iberoam. J. Med. 2 (2020) 322 (http://dx.doi.org/10.53986/ibjm.2020.0056)
N. Contreras-Puentes, M. Salas-Moreno, L. Mosquera-Chaverra, L. Córdoba-Tovar, A. Alviz-Amador, J. Pharm. Pharmacogn. Res. 10 (2022) 469 (http://dx.doi.org/10.56499/jppres21.1328_10.3.469)
P. T. Quy, T. Q. Bui, N. M. Thai, L. N. H. Du, N. T. Triet, T. Van Chen, N. V. Phu, D. T. Quang, D. C. To, N. T. A. Nhung, Open Chem. 21 (2023) 20230109 (http://dx.doi.org/10.1515/chem-2023-0109)
S. Dev, I. Kaur, Kragujev. J. Sci. 42 (2020) 29 (http://dx.doi.org/10.5937/kgjsci2042029d)
U. J. Meierhenrich, Rev. Oenologues Tech. Vitivinic. Oenologiques Mag. Trimest. Inform. 33 (2006) 19 (https://dialnet.unirioja.es/servlet/articulo?codigo=3556037)
V. Meyer, Détection d’homologies lointaines à faibles identités de séquences : Application aux protéines de la signalisation des dommages de l’ADN, Université Paris-
-Diderot – Paris VII, 2007 (https://theses.hal.science/tel-00361212)
I. Aanouz, A. Belhassan, K. El-Khatabi, T. Lakhlifi, M. El-ldrissi, M. Bouachrine, J. Biomol. Struct. Dyn. 39 (2021) 2971 (http://dx.doi.org/10.1080/07391102.2020.1758790)
H. Zaki, A. Belhassan, M. Benlyas, T. Lakhlifi, M. Bouachrine, J. Biomol. Struct. Dyn. 39 (2021) 2993 (http://dx.doi.org/10.1080/07391102.2020.1759452)
K. Arnold, L. Bordoli, J. Kopp, T. Schwede, Bioinformatics 22 (2006) 195 (http://dx.doi.org/10.1093/BIOINFORMATICS/BTI770)
A. Belhassan, H. Zaki, A. Aouidate, M. Benlyas, T. Lakhlifi, M. Bouachrine, Moroccan J. Chem. 7 (2019) 028 (http://dx.doi.org/10.48317/IMIST.PRSM/MORJCHEM-V7I1.12247)
A. Belhassan, S. Chtita, H. Zaki, T. Lakhlifi, M. Bouachrine, Bioinformation 16 (2020) 404 (http://dx.doi.org/10.6026/97320630016404)
M. Biasini, S. Bienert, A. Waterhouse, K. Arnold, G. Studer, T. Schmidt, F. Kiefer, T. G. Cassarino, M. Bertoni, L. Bordoli, T. Schwede, Nucleic Acids Res. 42 (2014) w252 (http://dx.doi.org/10.1093/NAR/GKU340)
N. Guex, M. C. Peitsch, T. Schwede, Electrophoresis 30 (2009) S162 (http://dx.doi.org/10.1002/ELPS.200900140)
H. E. Pence, A. Williams, J. Chem. Educ. 87 (2010) 1123 (http://dx.doi.org/10.1021/ED100697W)
L. Ahmed, Y. Zhang, E. Block, M. Buehl, M. J. Corr, R. A. Cormanich, S. Gundala, H. Matsunami, D. O’Hagan, M. Ozbil, Y. Pan, S. Sekharan, N. Ten, M. Wang, M. Yang, Q. Zhang, R. Zhang, V. S. Batista, H. Zhuang, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) E3950 (https://doi.org/10.1073/pnas.1713026115)
O. Trott, A. J. Olson, J. Comput. Chem. 31 (2010) 455 (http://dx.doi.org/10.1002/jcc.21334)
BIOVIA, Dassault Systèmes, Discovery Studio Visualiser 2019, San Diego: Dassault Systèmes, 2019 (https://discover.3ds.com/discovery-studio-visualizer-download)
H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, P. E. Bourne, Nucleic Acids Res. 28 (2000) 235 (http://dx.doi.org/10.1093/nar/28.1.235)
M. Hakmi, E. M. Bouricha, I. Kandoussi, J. El Harti, A. Ibrahimi, Bioinformation 16 (2020) 301 (http://dx.doi.org/10.6026/97320630016301)
C. Kutzner, S. Páll, M. Fechner, A. Esztermann, B. L. De Groot, H. Grubmüller, J. Comput. Chem. 36 (2015) 1990 (http://dx.doi.org/10.1002/JCC.24030)
E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, J. Comput. Chem. 25 (2004) 1605 (http://dx.doi.org/10.1002/jcc.20084)
M. Martínez-Cifuentes, B. E. Weiss-López, L. S. Santos, R. Araya-Maturana, Molecules 19 (2014) 9354 (http://dx.doi.org/10.3390/MOLECULES19079354)
A. Kumar, C. G. Mohan, P. C. Mishra, J. Mol. Struct. Theochem 361 (1996) 135 (http://dx.doi.org/10.1016/0166-1280(95)04312-8)
K. Raghavachari, Theor. Chem. Accounts 103 (2000) 361 (https://doi.org/10.1007/S002149900065)
A. D. Becke, Phys. Rev., A 38 (1988) 3098 (http://dx.doi.org/https://doi.org/10.1103/PhysRevA.38.3098)
Gaussian 09, Revision A.01, Gaussian, Inc., Wallingford, CT, 2009. (https://gaussian.com/g09citation/)
Gaussview Rev. 3.09, Windows version, Gaussian Inc., Pittsburgh, PA (https://gaussian.com/508_gvw/)
T. Pantsar, A. Poso, Molecules 23 (2018) 1899 (http://dx.doi.org/10.3390/molecules23081899)
M. Popovic, Microb. Risk Anal. 23 (2023) 100250 (http://dx.doi.org/10.1016/j.mran.2023.100250)
M. Popovic, Microb. Risk Anal. 22 (2022) 100231 (http://dx.doi.org/10.1016/j.mran.2022.100231)
X. Du, Y. Li, Y. L. Xia, S. M. Ai, J. Liang, P. Sang, X. L. Ji, S. Q. Liu, Int. J. Mol. Sci. 17 (2016) 144 (http://dx.doi.org/10.3390/ijms17020144)
P. Gale, Microb. Risk Anal. 21 (2022) 100198 (http://dx.doi.org/10.1016/j.mran.2021.100198)
T. Maass, G. Ssebyatika, M. Brückner, L. Breckwoldt, T. Krey, A. Mallagaray, T. Peters, M. Frank, R. Creutznacher, Chem. - A Eur. J. 28 (2022) e202202614 (http://dx.doi.org/10.1002/chem.202202614).