GC–MS-based metabolomics for the detection of adulteration in oregano samples Scientific paper
Main Article Content
Abstract
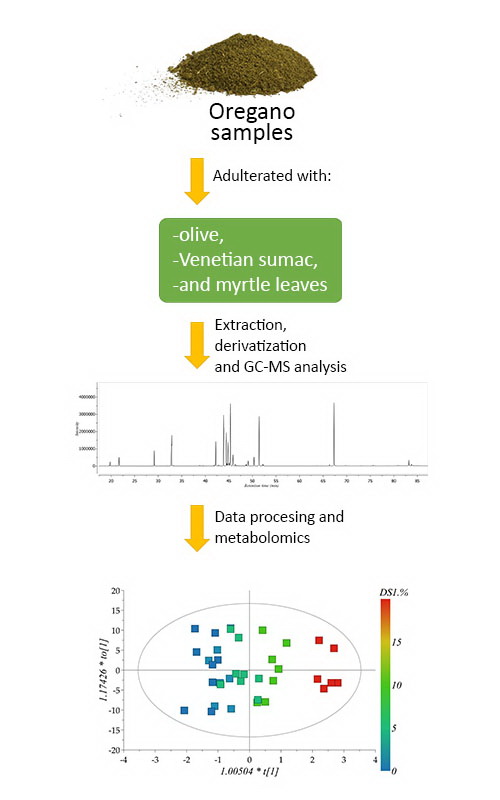
Oregano is one of the most used culinary herb and it is often adulterated with cheaper plants. In this study, GC–MS was used for identification and quantification of metabolites from 104 samples of oregano (Origanum vulgare and O. onites) adulterated with olive (Olea europaea), venetian sumac (Cotinus coggygria) and myrtle (Myrtus communis) leaves, at five different concentration levels. The metabolomics profiles obtained after the two-step derivatization, involving methoxyamination and silanization, were subjected to multivariate data analysis to reveal markers of adulteration and to build the regression models on the basis of the oregano-to-adulterants mixing ratio. Orthogonal partial least squares enabled detection of oregano adulterations with olive, Venetian sumac and myrtle leaves. Sorbitol levels distinguished oregano samples adulterated with olive leaves, while shikimic and quinic acids were recognized as discrimination factor for adulteration of oregano with venetian sumac. Fructose and quinic acid levels correlated with oregano adulteration with myrtle. Orthogonal partial least squares discriminant analysis enabled discrimination of O. vulgare and O. onites samples, where catechollactate was found to be discriminating metabolite.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
P. Galvin-King, S. A. Haughey, C. T. Elliott, Food Control 88 (2018) 85 (https://doi.org/10.1016/j.foodcont.2017.12.031)
S. Lohumi, S. Lee, H. Lee, B.-K. Cho, Trends Food Sci. Technol. 46 (2015) 85 (https://doi.org/10.1016/j.tifs.2015.08.003)
C. Black, S. A. Haughey, O. P. Chevallier, P. Galvin-King, C. T. Elliott, Food Chem. 210 (2016) 551 (https://doi.org/10.1016/j.foodchem.2016.05.004)
L. Drabova, G. Alvarez-Rivera, M. Suchanova, D. Schusterova, J. Pulkrabova, M. Tomaniova, V. Kocourek, O. Chevallier, C. Elliott, J. Hajslova, Food Chem. 276 (2019) 726 (https://doi.org/10.1016/j.foodchem.2018.09.143)
M. Marieschi, A. Torelli, A. Bianchi, R. Bruni, Food Control 22 (2011) 542
(https://doi.org/10.1016/j.foodcont.2010.10.001)
M. Mandrone, L. Marincich, I. Chiocchio, A. Petroli, D. Gođevac, I. Maresca, F. Poli, Food Control 127 (2021) 108141. (https://doi.org/10.1016/j.foodcont.2021.108141)
L. E. Rodriguez-Saona, M. E. Allendorf, Ann. Rev. Food Sci. Technol. 2 (2011) 467 (https://doi.org/10.1146/annurev-food-022510-133750)
M. Bononi, I. Fiordaliso, F. Tateo, Ital. J. Food Sci. 22 (2010) 479 (https://www.researchgate.net/publication/288447203_Rapid_GCMS_test_for_identification_of_Olea_Europaea_L_leaves_in_ground_oregano)
P. Galvin-King, S. A. Haughey, C. T. Elliott, Food Control 88 (2018) 85 (https://doi.org/10.1016/j.foodcont.2017.12.031)
European Directorate for the Quality of Medicines & HealthCare, European Pharmacopoeia, 9th ed., European Pharmacopoeia, Strasbourg, 2017
J. H. Ietswaart, A Taxonomie Revision of the Genus Origanum (Labiatae), Leiden University Press, The Hague, 1982 (https://doi.org/10.1002/fedr.19820930120)
J. Lisec, N. Schauer, J. Kopka, L. Willmitzer, A. R. Fernie, Nat. Protoc. 1 (2006) 387 (https://doi.org/10.1038/nprot.2006.59)
E. Dervishi, G. Zhang, G. Zwierzchowski, R. Mandal, D. S. Wishart, B. N. Ametaj, J. Proteomics 213 (2020) 103620 (https://doi.org/10.1016/j.jprot.2019.103620)
C. A. Smith, E. J. Want, G. O’Maille, R. Abagyan, G. Siuzdak, Anal. Chem. 78 (2006) 779 (https://doi.org/10.1021/ac051437y)
R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, 2018 (https://www.R-project.org/)
S. Wiklund, E. Johansson, L. Sjöström, E. J. Mellerowicz, U. Edlund, J. P. Shockcor, J. Gottfries, T. Moritz, J. Trygg, Anal. Chem. 80 (2008) 115 (https://doi.org/10.1021/ac0713510).