A computational study of the potential bioactivity of hibiscus and garcinia acids against SARS-CoV-2 Scientific paper
Main Article Content
Abstract
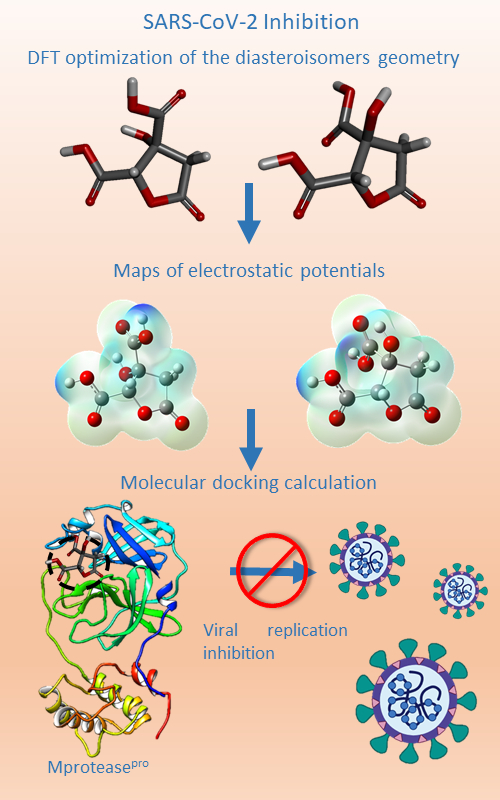
A computational chemical study was conducted on the diastereoisomers of hibiscus acid (HA) and garcinia acid (GA), investigating their docking capabilities with the main protease (6LU7) of SARS-CoV-2. Electrostatic potential mappings unveiled negative charges associated with the carboxyl and hydroxyl groups positioned at C-2 and C-3 for both hibiscus and garcinia acids. However, the presence of more negative potentials around C-2 and C-3 of hibiscus acid, compared to garcinia acid, suggests that substituents in the (2S,3R) configuration possess a stronger electron-attracting capacity than those in the (2S,3S) configuration. Molecular docking studies indicated that both hibiscus acid and garcinia acid bind to the main protease through the catalytic pocket. Nonetheless, molecular dynamics simulations revealed that only HA remained bound to the active site for 100 ns with an RMSD of less than 1 Å, whereas GA dissociated from the complex within the initial 16 ns. These findings illuminate the differential binding behaviors of the two compounds, with implications for potential therapeutic interventions against SARS-CoV-2. These findings shed light on the differential binding behaviors of the two compounds, holding implications for potential therapeutic interventions against SARS-CoV-2.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Universidad Autónoma del Estado de Hidalgo
Grant numbers PAO-2022-1389 -
Consejo Nacional de Ciencia y Tecnología
Grant numbers CB2015-257823
References
T. I. Ng, I. Correia, J. Seagal, D. A. Degoey, M. R. Schrimpf, D. J. Hardee, E. L. Noey, W. M. Kati, Viruses 14 (2022) 961 (http://dx.doi.org/10.3390/V14050961)
Y. L. Ng, C. K. Salim, J. J. H. Chu, Pharmacol. Ther. 228 (2021) 107930 (http://dx.doi.org/10.1016/J.PHARMTHERA.2021.107930)
C. L. Bellera, M. Llanos, M. E. Gantner, S. Rodriguez, L. Gavernet, M. Comini, A. Talevi, Expert Opin. Drug Discov. 16 (2021) 605 (http://dx.doi.org/10.1080/17460441.2021.1863943)
I. Aanouz, A. Belhassan, K. El-Khatabi, T. Lakhlifi, M. El-ldrissi, M. Bouachrine, J. Biomol. Struct. Dyn. 39 (2021) 2971 (http://dx.doi.org/10.1080/07391102.2020.1758790)
S. Yagi, A. Yagi, Curr. Tradit. Med. 9 (2023) 39 (http://dx.doi.org/10.2174/2215083809666230206114117)
A. Ghosh, S. Chakraborty, S. Majumder, M. Bhattacharya, Lett. Appl. NanoBioScience 12 (2023) 26 (http://dx.doi.org/10.33263/LIANBS124.108)
M. T. Quimque, K. I. Notarte, X. A. Adviento, M. H. Cabunoc, V. N. de Leon, F. S. L. delos Reyes, E. J. Lugtu, J. A. Manzano, S. N. Monton, J. E. Muñoz, K. D. Ong, D. Y. Pilapil, V. Roque, S. M. Tan, J. A. Lim, A. P. Macabeo, Comb. Chem. High Throughput Screen. 26 (2021) 459 (http://dx.doi.org/10.2174/1386207325666210917113207)
R. B. Malabadi, K. P. Kolkar, N. T. Meti, R. K. Chalannavar, Int. J. Innov. Sci. Res. Rev. 3 (2021) 1507 (https://journalijisr.com/sites/default/files/issues-pdf/IJISRR-600.pdf)
Y. T. M. Alharbi, W. M. Abdel-Mageed, O. A. Basudan, R. A. Mothana, M. Tabish Rehman, A. A. ElGamal, A. S. Alqahtani, O. I. Fantoukh, M. F. AlAjmi, Saudi Pharm. J. 32 (2024) 102023 (http://dx.doi.org/10.1016/J.JSPS.2024.102023)
M. Ariefin, R. R. Saputra, I. N. Pramesti, AIP Conf. Proc. 3055 (2024) 040001 (https://doi.org/10.1063/5.0193646)
S. T. Selvan, M. K. D. Jothinathan, Cureus 16 (2024) 1 (http://dx.doi.org/10.7759/CUREUS.57151)
M. A. Khanfar, M. I. Saleh, Curr. Med. Chem. 16 (2024) e57151 (http://dx.doi.org/10.2174/0109298673271674231109052709)
E. Akbaba, D. Karatas, J. Inst. Sci. Tech. 13 (2023) 872 (http://dx.doi.org/10.21597/jist.1187616)
S. Das, S. Satapathy, D. Acharya, S. Kumar Sahu, Res. Sq. 2023 (2023) 1 (http://dx.doi.org/10.21203/RS.3.RS-2837087/V1)
C. Parga-Lozano, Duazary 17 (2020) 1 (http://dx.doi.org/10.21676/2389783x.3597)
C. H. Parga-Lozano, Biomed. J. Sci. Tech. Res. 35 (2021) 28000 (http://dx.doi.org/10.26717/bjstr.2021.35.005761)
N. Balmeh, S. Mahmoudi, N. Mohammadi, A. Karabedianhajiabadi, Informatics Med. Unlocked 20 (2020) 100407 (http://dx.doi.org/10.1016/J.IMU.2020.100407)
S. Mahmoudi, N. Balmeh, N. Mohammadi, T. Sadeghian-Rizi, Avicenna J. Med. Biotechnol. 13 (2021) 107 (http://dx.doi.org/10.18502/AJMB.V13I3.6370)
N. Ohta, K. Inokuma, K. Miyabayashi, M. Miyake, I. Yagi, Electrochemistry 78 (2012) 132 (http://dx.doi.org/10.5796/electrochemistry.78.132)
N. F. Ramadhani, A. P. Nugraha, D. Rahmadhani, M. S. Puspitaningrum, Y. Rizqianti, V. D. Kharisma, T. N. E. B. T. A. Noor, R. D. Ridwan, D. S. Ernawati, A. P. Nugraha, J. Pharm. Pharmacogn. Res. 10 (2022) 418 (http://dx.doi.org/10.56499/jppres21.1316_10.3.418)
S. Guardiola, N. Mach, Endocrinol. y Nutr. 61 (2014) 274 (http://dx.doi.org/10.1016/j.endonu.2013.10.012)
E. Shawky, A. A. Nada, R. S. Ibrahim, RSC Adv. 10 (2020) 27961 (http://dx.doi.org/10.1039/d0ra05126h)
O. A. Olajide, V. U. Iwuanyanwu, I. Lepiarz-Raba, A. A. Al-Hindawi, M. A. Aderogba, H. L. Sharp, R. J. Nash, Phyther. Res. 35 (2021) 6963 (http://dx.doi.org/10.1002/ptr.7315)
H. Y. Aati, A. Ismail, M. E. Rateb, A. M. AboulMagd, H. M. Hassan, M. H. Hetta, Plants 11 (2022) 2521 (http://dx.doi.org/10.3390/PLANTS11192521)
A. J. Akindele, A. Sowemimo, F. O. Agunbiade, M. O. Sofidiya, O. Awodele, O. Ade-Ademilua, I. Orabueze, I. O. Ishola, C. I. Ayolabi, O. B. Salu, M. O. Akinleye, I. A. Oreagba, Nat. Prod. Commun. 17 (2022) 1 (https://doi.org/10.1177/1934578X221096968)
O. P. Abodunrin, O. F. Onifade, A. E. Adegboyega, Informatics Med. Unlocked 31 (2022) 100964 (http://dx.doi.org/10.1016/J.IMU.2022.100964)
D. Muralitharan, V. Varadharajan, B. Venkidasamy, J. Mol. Recognit. 36 (2023) e3055 (http://dx.doi.org/10.1002/JMR.3055)
N. S. Aini, V. D. Kharisma, M. H. Widyananda, A. A. A. Murtadlo, R. T. Probojati, D. D. R. Turista, M. B. Tamam, V. Jakhmola, E. Yuniarti, S. Al Aziz, M. R. Ghifari, M. T. Albari, R. S. Mandeli, M. A. Ghifari, D. Purnamasari, B. Oktavia, A. P. Lubis, F. Azra, F. Fitri, A. N. M. Ansori, M. Rebezov, R. Zainul, Pharmacogn. J. 14 (2022) 575 (http://dx.doi.org/10.5530/pj.2022.14.138)
A. N. M. Ansori, V. D. Kharisma, A. A. Parikesit, F. A. Dian, R. T. Probojati, M. Rebezov, P. Scherbakov, P. Burkov, G. Zhdanova, A. Mikhalev, Y. Antonius, M. R. F. Pratama, N. I. Sumantri, T. H. Sucipto, R. Zainul, Pharmacogn. J. 14 (2022) 85 (http://dx.doi.org/10.5530/pj.2022.14.12)
V. D. Kharisma, A. N. M. Ansori, Y. Antonius, I. Rosadi, A. A. A. Murtadlo, V. Jakhmola, M. Rebezov, N. Maksimiuk, E. Kolesnik, P. Burkov, M. Derkho, P. Scherbakov, M. E. Ullah, T. H. Sucipto, H. Purnobasuki, J. Pharm. Pharmacogn. Res. 11 (2023) 743 (http://dx.doi.org/10.56499/JPPRES23.1650_11.5.743)
O. A. Kolawole, T. G. Femi, O. E. Kolawole, O. O. Monisola, O. Temitope, A. B. Benjamin, A. S. Adewale, S. Banj, Nat. Sci. 18 (2020) 78 (http://dx.doi.org/10.7537/marsnsj180920.10)
A. Kalita, M. Das, B. Das, M. R. Baro, Beni-Suef Univ. J. Basic Appl. Sci. 11 (2022) 1 (http://dx.doi.org/10.1186/S43088-022-00214-2/FIGURES/10)
N. Khamthong, N. Hutadilok-Towatana, Nat. Prod. Commun. 12 (2017) 453 (http://dx.doi.org/10.1177/1934578x1701200337)
G. I. P. Pozos, M. A. Ruiz-López, J. F. Z. Nátera, C. Á. Moya, L. B. Ramírez, M. R. Silva, R. R. Macías, P. M. García-López, R. G. Cruz, E. S. Pérez, J. J. V. Radillo, Appl. Sci. 10 (2020) 560 (http://dx.doi.org/10.3390/app10020560)
P. M. Afladhanti, M. D. Romadhan, H. A. Hamzah, Q. Bhelqis, Sriwij. J. Med. 5 (2022) 31 (http://dx.doi.org/10.32539/SJM.V5I1.127)
A. N. M. Ansori, V. D. Kharisma, A. A. Parikesit, F. A. Dian, R. T. Probojati, M. Rebezov, P. Scherbakov, P. Burkov, G. Zhdanova, A. Mikhalev, Y. Antonius, M. R. F. Pratama, N. I. Sumantri, T. H. Sucipto, R. Zainul, Pharmacogn. J. 14 (2022) 85 (http://dx.doi.org/10.5530/PJ.2022.14.12)
D. Ganjewala, H. Bansal, R. Mittal, G. Srivastava, Herb. Med. A Boon Heal. Hum. Life (2022) 471 (http://dx.doi.org/10.1016/B978-0-323-90572-5.00012-3)
J. A. Izquierdo-Vega, D. A. Arteaga-Badillo, M. Sánchez-Gutiérrez, J. A. Morales-Gon¬zález, N. Vargas-Mendoza, C. A. Gómez-Aldapa, J. Castro-Rosas, L. Delgado-Olivares, E. Madrigal-Bujaidar, E. Madrigal-Santillán, Biomedicines 8 (2020) 100 (http://dx.doi.org/10.3390/BIOMEDICINES8050100)
R. A. El-Shiekh, U. R. Abdelmohsen, H. M. Ashour, R. M. Ashour, Antibiotics 9 (2020) 756 (http://dx.doi.org/10.3390/antibiotics9110756)
L. O. Chuah, W. Y. Ho, B. K. Beh, S. K. Yeap, Evidence-Based Complement. Altern. Med. 2013 (2013) 751658 (http://dx.doi.org/10.1155/2013/751658)
S. Kim, Y. Kim, J. W. Kim, Y. Hwang, S. H. Kim, Y. H. Jang, J. Life Sci. 32 (2022) 375 (https://doi.org/10.5352/JLS.2022.32.5.375)
Y. Takeda, Y. Okuyama, H. Nakano, Y. Yaoita, K. Machida, H. Ogawa, K. Imai, Food Environ. Virol. 12 (2020) 9 (http://dx.doi.org/10.1007/s12560-019-09408-x)
R. Krishnan, J. S. Binkley, R. Seeger, J. A. Pople, J. Chem. Phys. 72 (1980) 650 (http://dx.doi.org/10.1063/1.438955)
A. Grosdidier, V. Zoete, O. Michielin, Nucleic Acids Res. 39 (2011) W270 (http://dx.doi.org/10.1093/NAR/GKR366)
E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, J. Comput. Chem. 25 (2004) 1605 (http://dx.doi.org/10.1002/jcc.20084)
Biovia Dassault Systèmes, Discovery Studio Visualiser 2019, Dassault Systèmes, San Diego, CA, 2020 (http://dx.doi.org/https://discover.3ds.com/discovery-studio-visualizer-download)
C. Kutzner, S. Páll, M. Fechner, A. Esztermann, B. L. De Groot, H. Grubmüller, J. Comput. Chem. 36 (2015) 1990 (http://dx.doi.org/10.1002/JCC.24030)
J. V. Vermaas, D. J. Hardy, J. E. Stone, E. Tajkhorshid, A. Kohlmeyer, J. Chem. Inf. Model. 56 (2016) 1112 (http://dx.doi.org/10.1021/acs.jcim.6b00103)
W. L. Jorgensen, D. S. Maxwell, J. Tirado-Rives, J. Am. Chem. Soc. 118 (1996) 11225 (http://dx.doi.org/10.1021/ja9621760)
W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey, M. L. Klein, J. Chem. Phys. 79 (1983) 926 (http://dx.doi.org/10.1063/1.445869)
G. Xiong, Z. Wu, J. Yi, L. Fu, Z. Yang, C. Hsieh, M. Yin, X. Zeng, C. Wu, A. Lu, X. Chen, T. Hou, D. Cao, Nucleic Acids Res. 49 (2021) W5 (http://dx.doi.org/10.1093/nar/gkab255)
C. A. Lipinski, Drug Discov. Today Technol. 1 (2004) 337 (http://dx.doi.org/10.1016/J.DDTEC.2004.11.007)
J. L. Dahlin, M. A. Walters, Assay Drug Dev. Technol. 14 (2016) 168 (http://dx.doi.org/10.1089/adt.2015.674)
J. J. Irwin, K. G. Tang, J. Young, C. Dandarchuluun, B. R. Wong, M. Khurelbaatar, Y. S. Moroz, J. Mayfield, R. A. Sayle, J. Chem. Inf. Model. 60 (2020) 6065 (http://dx.doi.org/10.1021/ACS.JCIM.0C00675)
M. Sargolzaei, J. Mol. Graph. Model. 103 (2021) 107803 (http://dx.doi.org/10.1016/j.jmgm.2020.107803)
M. A. Said, D. J. O. Khan, F. F. Al-Blewi, N. S. Al-Kaff, A. A. Ali, N. Rezki, M. R. Aouad, M. Hagar, Vaccines 9 (2021) 1012 (https://doi.org/10.3390/vaccines9091012)
A. Marques Da Fonseca, A. Luthierre, G. Cavalcante, R. Mateus, M. Carvalho, J. Falcão Do Amaral, R. P. Colares, E. S. Marinho, M. M. Neto, P. Colares, Int. J. Res. -GRANTHAALAYAH 8 (2020) 164 (http://dx.doi.org/10.29121/GRANTHAALAYAH.V8.I11.2020.2342)
P. Gale, Microb. Risk Anal. 21 (2022) 100198 (http://dx.doi.org/10.1016/j.mran.2021.100198)
M. Popovic, Vaccines 10 (2022) 2112 (http://dx.doi.org/10.3390/vaccines10122112)
M. Popovic, J. H. Martin, R. J. Head, Heliyon 9 (2023) e17174 (http://dx.doi.org/10.1016/j.heliyon.2023.e17174)
P. Gale, Microb. Risk Anal. 16 (2020) 100140 (http://dx.doi.org/10.1016/j.mran.2020.100140)
M. E. Popović, G. Šekularac, M. Popović, Microb. Risk Anal. 26 (2024) 100290 (http://dx.doi.org/10.1016/J.MRAN.2024.100290)
J. M. Casasnovas, T. A. Springer, J. Biol. Chem. 270 (1995) 13216 (http://dx.doi.org/10.1074/jbc.270.22.13216)
Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao, Nature 582 (2020) 289 (http://dx.doi.org/10.1038/S41586-020-2223-Y).