π–π interactions in structural stability: Role in superoxide dismutases Scientific paper
Main Article Content
Abstract
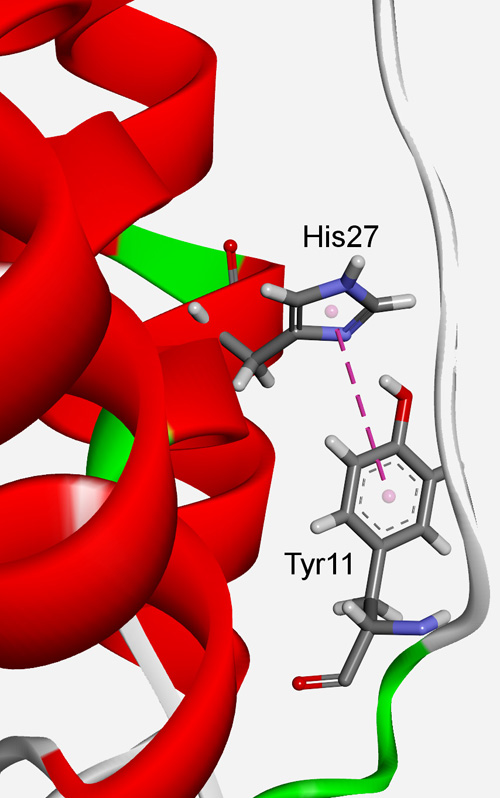
In the present work, the influences of π–π interactions in superoxide dismutase (SOD) active centers were analyzed. The majority of the aromatic residues are involved in π–π interactions. Predominant type of interacting pairs is His–His and His–Trp pairs. In addition to π–π interactions, π residues also form π-networks in SOD proteins. The π–π interactions are most favorable at the pair distance range of 5–7 Å. We observed that most of the π–π interactions shows stabilization energies in the range from −4.2 to −12.6 kJ mol-1, while the metal assisted π–π interactions showed an energy in the range from −83.7 to −334.7 kJ mol-1. Most of the π–π interacting residues were evolutionary conserved and thus probably important in maintaining the structural stability of proteins through these interactions. A high percentage of these residues could be considered as stabilization centers, contributing to the net stability of SOD proteins.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-68/2022-14/200026 and 451-03-68/2022-14/200168
References
C. D. Andersson, B. K. Mishra, N. Forsgren, F. Ekström, A. Linusson, J. Phys. Chem., B 124 (2020) 6529 (https://doi.org/10.1021/acs.jpcb.0c03778)
E. Lanzarotti, L. A. Defelipe, M. A. Marti, A. n. G. Turjanski, J. Cheminf. 12 (2020) 30 (https://doi.org/10.1186/s13321-020-00437-4)
K. S. Chatterjee, R. Das, J. Biol. Chem. 297 (2021) (https://doi.org/10.1016/j.jbc.2021.100970)
H. B. Gray, J. R. Winkler, Chem. Sci. 12 (2021) 13988 (https://doi.org/10.1039/D1SC04286F)
Z. Y. Yan, X. J. Xu, L. Fang, C. Geng, Y. P. Tian, X. D. Li, Phytopathol. Res. 3 (2021) 10 (https://doi.org/10.1186/s42483-021-00088-9)
S. Sasidharan, V. Ramakrishnan, Aromatic interactions directing peptide nano-assembly. Advances in Protein Chemistry and Structural Biology. Academic Press, Cambridge, MA, 2022 (https://doi.org/10.1016/bs.apcsb.2022.01.001)
S. K. Burley, G. A. Petsko, Science 229 (1985) 23 (https://www.science.org/doi/10.1126/science.8235619)
E. Cauët, M. Rooman, R. Wintjens, J. Liévin, C. Biot, J. Chem. Theory Comput. 1 (2005) 472 (https://doi.org/10.1021/ct049875k)
M. O. Sinnokrot, C. D. Sherrill, J. Am. Chem. Soc. 126 (2004) 7690 (https://doi.org/10.1021/ja049434a)
G. B. McGaughey, M. Gagné, A. K. Rappé, J. Biol. Chem. 273 (1998) 15458 (https://doi.org/10.1074/jbc.273.25.15458)
R. Bhattacharyya, U. Samanta, P. Chakrabarti, Protein Eng. 15 (2002) 91 (https://doi.org/10.1093/protein/15.2.91)
N. Kannan, S. Vishveshwara, Protein Eng. 13 (2000) 753 (https://doi.org/10.1093/protein/13.11.753)
S. Tsuzuki, K. Honda, T. Uchimaru, M. Mikami, K. Tanabe, J. Am. Chem. Soc. 124 (2002) 104 (https://doi.org/10.1021/ja0105212)
A. V. Morozov, K. M. S. Misura, K. Tsemekhman, D. Baker, J. Phys. Chem., B 108 (2004) 8489 (https://doi.org/10.1021/jp037711e)
C. Chipot, R. Jaffe, B. Maigret, D. A. Pearlman, P. A. Kollman, J. Am. Chem. Soc. 118 (1996) 11217 (https://doi.org/10.1021/ja961379l)
E. Lanzarotti, R. R. Biekofsky, D. O. A. Estrin, M. A. Marti, A. n. G. Turjanski, J. Chem. Inf. Model. 51 (2011) 1623 (https://doi.org/10.1021/ci200062e)
V. R. Ribić, S. Đ. Stojanović, M. V. Zlatović, Int. J. Biol. Macromol. 106 (2018) 559 (https://doi.org/10.1016/j.ijbiomac.2017.08.050)
S. Stojanović, M. Zlatović, J. Serb. Chem. Soc. 87 (2022) 465 (https://doi.org/10.2298/JSC220109013S)
P. W. Rose, B. Beran, C. Bi, W. F. Bluhm, D. Dimitropoulos, D. S. Goodsell, A. Prlic, M. Quesada, G. B. Quinn, J. D. Westbrook, J. Young, B. Yukich, C. Zardecki, H. M. Berman, P. E. Bourne, Nucleic Acids Res. 39 (2011) D392 (https://doi.org/10.1093/nar/gkq1021
J. M. Word, S. C. Lovell, J. S. Richardson, D. C. Richardson, J. Mol. Biol. 285 (1999) 1735 (https://doi.org/10.1006/jmbi.1998.2401)
Discovery Studio Visualizer, Release 2020. Accelrys Software Inc., San Diego, CA
J. Hostaš, D. Jakubec, R. A. Laskowski, R. Gnanasekaran, J. Řezáč, J. Vondrášek, P. Hobza, J. Chem. Theory. Comput. 11 (2015) 4086 (http://dx.doi.org/10.1021/acs.jctc.5b00398)
Schrödinger, Release 2018-1: Jaguar, Schrödinger, LLC, New York, 2018
T. H. Dunning, J. Chem. Phys. 90 (1989) 1007 (https://doi.org/10.1063/1.456153)
T. Clark, J. Chandrasekhar, G. n. W. Spitznagel, P. V. R. Schleyer, J. Comput. Chem. 4 (1983) 294 (https://doi.org/10.1002/jcc.540040303)
A. D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang, R. A. Friesner, Int. J. Quantum Chem. 113 (2013) 2110 (https://doi.org/10.1002/qua.24481)
K. E. Riley, J. A. Platts, J. Řezáč, P. Hobza, J. G. Hill, J. Phys. Chem., A 116 (2012) 4159 (https://doi.org/10.1021/jp211997b)
G. J. Jones, A. Robertazzi, J. A. Platts, J. Phys. Chem., B 117 (2013) 3315 (https://doi.org/10.1021/jp400345s)
S. Saebø, W. Tong, P. Pulay, J. Chem. Phys. 98 (1993) 2170 (https://doi.org/10.1063/1.464195)
A. Reyes, L. Fomina, L. Rumsh, S. Fomine, Int. J. Quantum Chem. 104 (2005) 335 (https://doi.org/10.1002/qua.20558)
R. M. Balabin, J. Chem. Phys. 132 (2010) 231101 (https://doi.org/10.1063/1.3442466)
P. J. Hay, W. R. Wadt, J. Chem. Phys. 82 (1985) 299 (https://doi.org/10.1063/1.448975)
D. Vijay, G. N. Sastry, Chem. Phys. Lett. 485 (2010) 235 (https://doi.org/10.1016/j.cplett.2009.12.012)
Z. Dosztányi, A. Fiser, I. Simon, J. Mol. Biol. 272 (1997) 597 (https://doi.org/10.1006/jmbi.1997.1242)
Z. Dosztányi, C. Magyar, G. Tusnady, I. Simon, Bioinformatics 19 (2003) 899 (https://doi.org/10.1093/bioinformatics/btg110)
H. Ashkenazy, E. Erez, E. Martz, T. Pupko, N. Ben-Tal, Nucleic Acids Res. 38 (2010) W529 (https://doi.org/10.1093/nar/gkq399)
B. Boeckmann, A. Bairoch, R. Apweiler, M. C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O'Donovan, I. Phan, S. Pilbout, M. Schneider, Nucleic Acids Res. 31 (2003) 365 (https://doi.org/10.1093/nar/gkg095)
S. Stojanović, Z. Petrović, M. Zlatović, J. Serb. Chem. Soc. 86 (2021) 781 (https://doi.org/10.2298/JSC210321042S)
A. S. Mahadevi, G. N. Sastry, Chem. Rev. 116 (2016) 2775 (https://doi.org/10.1021/cr500344e)
B. Ma, T. Elkayam, H. Wolfson, R. Nussinov, Proc. Natl. Acad. Sci. USA 100 (2003) 5772 (https://doi.org/10.1073/pnas.1030237100)
E. G. Hohenstein, C. D. Sherrill, J. Phys. Chem., A 113 (2009) 878 (https://doi.org/10.1021/jp809062x)
P. Chakrabarti, R. Bhattacharyya, Prog. Biophys. Mol. Biol. 95 (2007) 83 (https://doi.org/10.1016/j.pbiomolbio.2007.03.016)
S. Marsili, R. Chelli, V. Schettino, P. Procacci, Phys. Chem. Chem. Phys. 10 (2008) 2673 (https://doi.org/10.1039/B718519G)
S. Ishikawa, T. Ebata, H. Ishikawa, T. Inoue, N. Mikami, J. Phys. Chem. 100 (1996) 10531 (https://doi.org/10.1021/jp960267d)
A. Banerjee, A. Saha, B. K. Saha, Crystal Growth Design 19 (2019) 2245 (https://doi.org/10.1021/acs.cgd.8b01857)
M. Landau, I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, N. Ben-Tal, Nucleic Acids Res. 33 (2005) W299-W302 (https://doi.org/10.1093/nar/gki370)