Amide–π interactions in active centers of superoxide dismutase Scientific paper
Main Article Content
Abstract
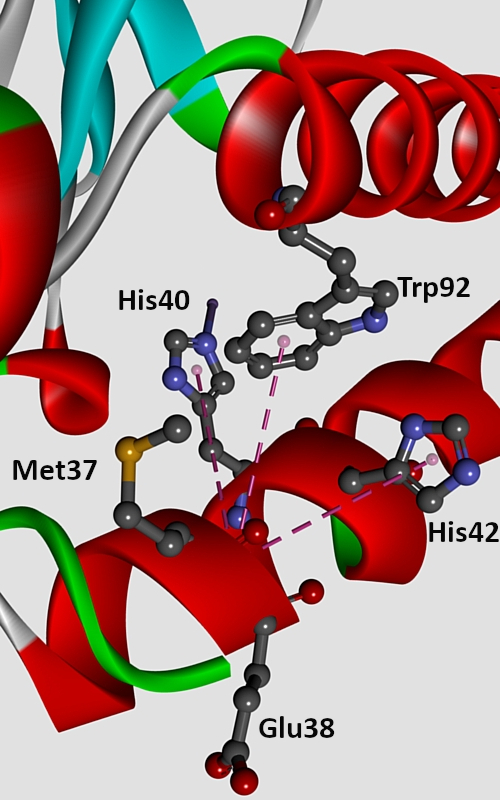
In this work, the influence of amide–π interactions on stability and properties of superoxide dismutase (SOD) active centres was analysed. In the data set of 43 proteins, 5017 amide–π interactions were observed, and every active centre formed averagely about 117 interactions. Most of the interactions belonged to the backbone of proteins. The analysis of the geometry of the amide–π interactions revealed two preferred structures, parallel-displaced and T-shaped structure. The aim of this study was to investigate the energy contribution resulting the from amide–π interactions, which were in the lower range of strong hydrogen bonds. The conservation patterns in the present study indicate that more than half of the residues involved in these interactions are evolutionarily conserved. The stabilization centres for these proteins showed that all residues involved in amide–π interactions were of use in locating one or more of such centres. The results presented in this work can be very useful for the understanding of contribution of amide–π interaction to the stability of SOD active centres.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
E. A. Meyer, R. K. Castellano, F. Diederich, Angew. Chem., Int. Ed. Engl. 42 (2003) 1210 (https://doi.org/10.1002/anie.200390319)
L. M. Salonen, M. Ellermann, F. Diederich, Angew. Chem., Int. Ed. Engl. 50 (2011) 4808 (https://doi.org/10.1002/anie.201007560)
N. Acharjee, J. Serb. Chem. Soc. 85 (2020) 765 (https://doi.org/10.2298/JSC190914136A)
M. Levitt, M. F. Perutz, J. Mol. Biol. 201 (1988) 751 (https://doi.org/10.1016/0022-2836(88)90471-8)
J. Cheney, B. V. Cheney, W. G. Richards, Biochim. Biophys. Acta 954 (1988) 137 (https://doi.org/10.1016/0167-4838(88)90063-5)
G. Duan, V. H. Smith, D. F. Weaver, Chem. Phys. Lett. 310 (1999) 323 (https://doi.org/10.1016/S0009-2614(99)00804-0)
M. Harder, B. Kuhn, F. Diederich, ChemMedChem 8 (2013) 397 (https://doi.org/10.1002/cmdc.201200512)
S. Cotesta, M. Stahl, J. Mol. Model. 12 (2006) 436 (https://doi.org/10.1007/s00894-005-0067-x)
S. K. Burley, G. A. Petsko, Science 229 (1985) 23 (https://doi.org/10.1126/science.3892686)
S. K. Burley, G. A. Petsko, FEBS Lett. 203 (1986) 139 (https://doi.org/10.1016/0014-5793(86)80730-x)
S. K. Burley, G. A. Petsko, Adv. Protein Chem. 39 (1988) 125 (https://doi.org/10.1016/s0065-3233(08)60376-9)
T. Steiner, G. Koellner, J. Mol. Biol. 305 (2001) 535 (https://doi.org/10.1006/jmbi.2000.4301)
F. R. Ferreira de, M. Schapira, MedChemComm 8 (2017) 1970 (https://doi.org/10.1039/c7md00381a)
M. Giroud, J. Ivkovic, M. Martignoni, M. Fleuti, N. Trapp, W. Haap, A. Kuglstatter, J. Benz, B. Kuhn, T. Schirmeister, F. Diederich, ChemMedChem 12 (2017) 257 (https://doi.org/10.1002/cmdc.201600563)
S. Raghunathan, T. Jaganade, U. D. Priyakumar, Biophys. Rev. 12 (2020) 65 (https://doi.org/10.1007/s12551-020-00620-9)
M. Giroud, M. Harder, B. Kuhn, W. Haap, N. Trapp, W. B. Schweizer, T. Schirmeister, F. Diederich, ChemMedChem 11 (2016) 1042 (https://doi.org/10.1002/cmdc.201600132)
L. M. Salonen, M. C. Holland, P. S. Kaib, W. Haap, J. Benz, J. L. Mary, O. Kuster, W. B. Schweizer, D. W. Banner, F. Diederich, Chemistry 18 (2012) 213 (https://doi.org/10.1002/chem.201102571)
B. S. Lauber, L. A. Hardegger, K. A. Alam, B. A. Lund, O. Dumele, M. Harder, B. Kuhn, R. A. Engh, F. Diederich, Chemistry 22 (2016) 211 (https://doi.org/10.1002/chem.201503552)
V. Ehmke, E. Winkler, D. W. Banner, W. Haap, W. B. Schweizer, M. Rottmann, M. Kaiser, C. Freymond, T. Schirmeister, F. Diederich, ChemMedChem 8 (2013) 967 (https://doi.org/10.1002/cmdc.201300112)
G. R. De, E. Brodbeck-Persch, S. Bryson, N. B. Hentzen, M. Kaiser, E. F. Pai, R. L. Krauth-Siegel, F. Diederich, ChemMedChem 13 (2018) 957 (https://doi.org/10.1002/cmdc.201800067)
M. W. Krone, C. R. Travis, G. Y. Lee, H. J. Eckvahl, K. N. Houk, M. L. Waters, J. Am. Chem. Soc. 142 (2020) 17048 (https://doi.org/10.1021/jacs.0c06568)
K. DeFrees, M. T. Kemp, X. ElHilali-Pollard, X. Zhang, A. Mohamed, Y. Chen, A. R. Renslo, Org. Chem. Front. 6 (2019) 1749 (https://doi.org/10.1039/c9qo00342h)
R. Meurisse, R. Brasseur, A. Thomas, Proteins 54 (2004) 478 (https://doi.org/10.1002/prot.10582)
P. W. Rose, B. Beran, C. Bi, W. F. Bluhm, D. Dimitropoulos, D. S. Goodsell, A. Prlic, M. Quesada, G. B. Quinn, J. D. Westbrook, J. Young, B. Yukich, C. Zardecki, H. M. Berman, P. E. Bourne, Nucleic Acids Res. 39 (2011) D392 (https://doi.org/10.1093/nar/gkq1021)
J. M. Word, S. C. Lovell, J. S. Richardson, D. C. Richardson, J. Mol. Biol. 285 (1999) 1735 (https://doi.org/10.1006/jmbi.1998.2401)
Accelrys Software Inc., Discovery Studio Visualizer, Release 2020, Accelrys Software Inc., San Diego, CA, 2020
M. R. Jackson, R. Beahm, S. Duvvuru, C. Narasimhan, J. Wu, H. N. Wang, V. M. Philip, R. J. Hinde, E. E. Howell, J. Phys. Chem., B 111 (2007) 8242 (https://doi.org/10.1021/jp0661995)
V. Philip, J. Harris, R. Adams, D. Nguyen, J. Spiers, J. Baudry, E. E. Howell, R. J. Hinde, Biochemistry 50 (2011) 2939 (https://doi.org/10.1021/bi200066k)
V. R. Ribić, S. Đ. Stojanović, M. V. Zlatović, Int. J. Biol. Macromol. 106 (2018) 559 (https://doi.org/10.1016/j.ijbiomac.2017.08.050)
J. Hostaš, D. Jakubec, R. A. Laskowski, R. Gnanasekaran, J. Řezáč, J. Vondrášek, P. Hobza, J. Chem. Theory Comput. 11 (2015) 4086 (http://dx.doi.org/10.1021/acs.jctc.5b00398)
Schrödinger Release 2018-1, Jaguar, Schrödinger, LLC, New York, NY, 2018
T. H. Dunning, J. Chem. Phys. 90 (1989) 1007 (https://doi.org/10.1063/1.456153)
T. Clark, J. Chandrasekhar, G. n. W. Spitznagel, P. V. R. Schleyer, J. Comput. Chem. 4 (1983) 294 (https://doi.org/10.1002/jcc.540040303)
A. D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang, R. A. Friesner, Int. J. Quantum Chem. 113 (2013) 2110 (https://doi.org/10.1002/qua.24481)
G. J. Jones, A. Robertazzi, J. A. Platts, J. Phys. Chem., B 117 (2013) 3315 (https://doi.org/10.1021/jp400345s)
K. E. Riley, J. A. Platts, J. Řezáč, P. Hobza, J. G. Hill, J. Phys. Chem., A 116 (2012) 4159 (https://doi.org/10.1021/jp211997b)
S. Saebø, W. Tong, P. Pulay, J. Chem. Phys. 98 (1993) 2170 (https://doi.org/10.1063/1.464195)
A. Reyes, L. Fomina, L. Rumsh, S. Fomine, Int. J. Quantum Chem. 104 (2005) 335 (https://doi.org/10.1002/qua.20558)
R. M. Balabin, J. Chem. Phys. 132 (2010) 231101 (https://doi.org/10.1063/1.3442466)
Z. Dosztányi, A. Fiser, I. Simon, J. Mol. Biol. 272 (1997) 597 (https://doi.org/10.1006/jmbi.1997.1242)
Z. Dosztányi, C. Magyar, G. Tusnady, I. Simon, Bioinformatics 19 (2003) 899 (https://doi.org/10.1093/bioinformatics/btg110)
H. Ashkenazy, E. Erez, E. Martz, T. Pupko, N. Ben-Tal, Nucleic Acids Res. 38 (2010) W529 (https://doi.org/10.1093/nar/gkq399)
B. Boeckmann, A. Bairoch, R. Apweiler, M. C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O'Donovan, I. Phan, S. Pilbout, M. Schneider, Nucleic Acids Res. 31 (2003) 365 (https://doi.org/10.1093/nar/gkg095)
G. Toth, C. R. Watts, R. F. Murphy, S. Lovas, Proteins 43 (2001) 373 (https://doi.org/10.1002/prot.1050)
A. S. Mahadevi, G. N. Sastry, Chem. Rev. 116 (2016) 2775 (https://doi.org/10.1021/cr500344e)
G. B. McGaughey, M. Gagne, A. K. Rappe, J. Biol. Chem. 273 (1998) 15458 (https://doi.org/10.1074/jbc.273.25.15458)
B. P. Dimitrijević, S. Z. Borozan, S. Đ. Stojanović, RSC Adv. 2 (2012) 12963 (https://doi.org/10.1039/C2RA21937A)
G. R. Desiraju, T. Steiner, The Weak Hydrogen Bond, Oxford University Press, Oxford, 1999.